Stabilize:
Predictive Mutation Platform
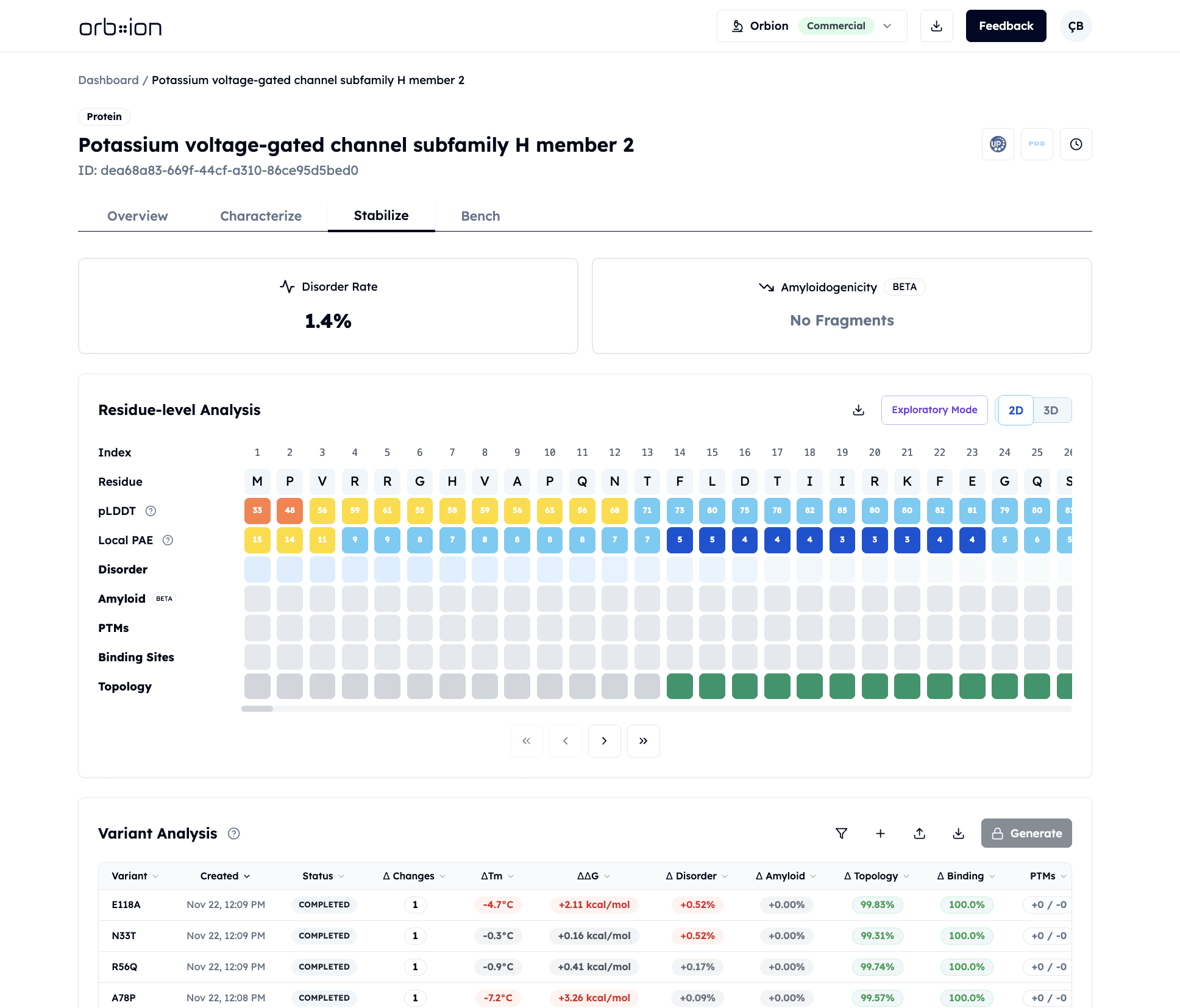
Stabilize transforms protein engineering from trial-and-error to rational design. Evaluate any mutation for stability changes, functional disruption, and aggregation risk—or generate optimized variants with improved Tm. Stabilize predicts melting temperature changes, free energy shifts, and checks for unintended consequences (PTM disruption, binding site damage, topology changes).
Protein engineering is expensive guesswork.
Designing stable proteins requires testing dozens of mutations to find the few that improve stability without breaking function. Each round costs weeks and €10K-50K. Mutations that increase Tm might disrupt binding sites, eliminate PTMs, or trigger aggregation—problems you discover after expression and purification.
Blind Mutation Testing
Rational design tools suggest mutations, but you don't know stability impact until wet lab testing. Each mutation requires expression, purification, and DSF/DSC—2-4 weeks and €5K-10K per variant.
Functional Damage
Stabilizing mutations can disrupt PTMs, binding sites, or critical interfaces. Surface entropy reduction removes a phosphorylation site. Thermostable mutation blocks ligand binding. Each functional failure wastes an entire experimental cycle.
Aggregation Risk
Some stabilizing mutations expose hydrophobic patches or create amyloidogenic sequences. Protein expression looks good, but aggregates form during purification or storage. Subtle sequence changes with catastrophic outcomes.
From Blind Testing to Predictive Design with Stabilize
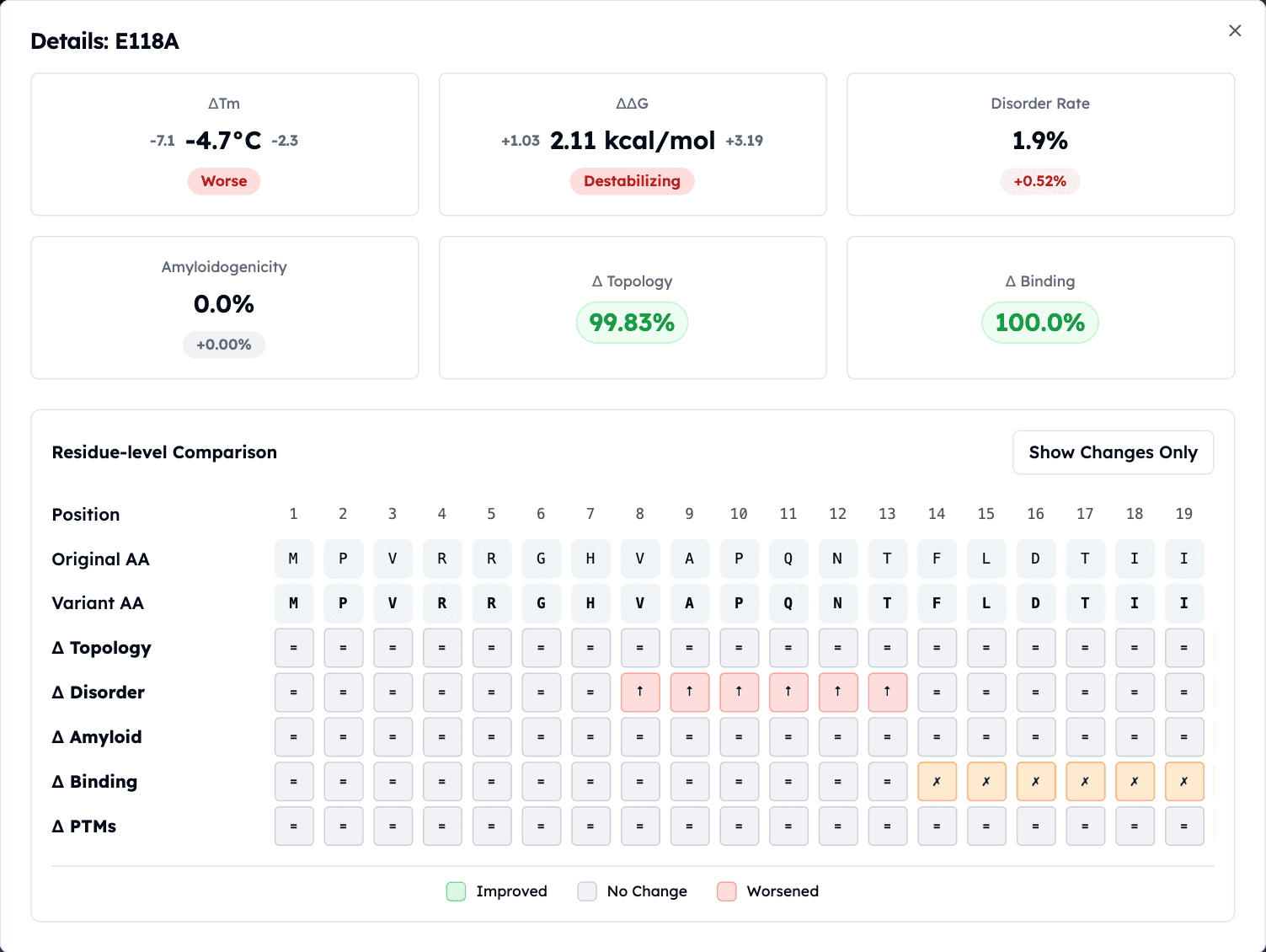
Stabilize predicts stability changes (ΔTm, ΔΔG) and checks for functional disruption before experiments. Evaluate your mutation ideas or generate optimized variants—then test only the candidates that improve stability without breaking function.
ΔTm and ΔΔG Visibility
Enter any mutation and get predicted melting temperature change (ΔTm) and free energy change (ΔΔG). AstraSTASIS predicts stability impact in seconds. Prioritize promising candidates before wet lab work.
Functional Preservation
Stabilize flags mutations that disrupt PTMs, binding sites, interfaces, or membrane topology. Design with guardrails. Improve stability while preserving function. No more discovering functional damage after purification.
AI-Generated Designs
Don't know where to start? Stabilize generates mutation combinations predicted to increase Tm by over 2°C. Surface entropy reduction, core packing, electrostatic optimization—all screened for functional safety.