Blog
The Tag Removal Problem: Why Your Protease Won't Cleave
Feb 4, 2026
The purification went perfectly. His-tag affinity, ion exchange, gel filtration—textbook chromatography. Now you just need to remove the tag before crystallization. You add TEV protease. Incubate overnight. Run the gel. Nothing happened. The band hasn't shifted. Your beautiful pure protein still has its tag, and your crystallization trials are on hold.
Tag removal should be straightforward: add protease, incubate, purify. In practice, it's one of the most frustrating steps in recombinant protein production.
Key Takeaways
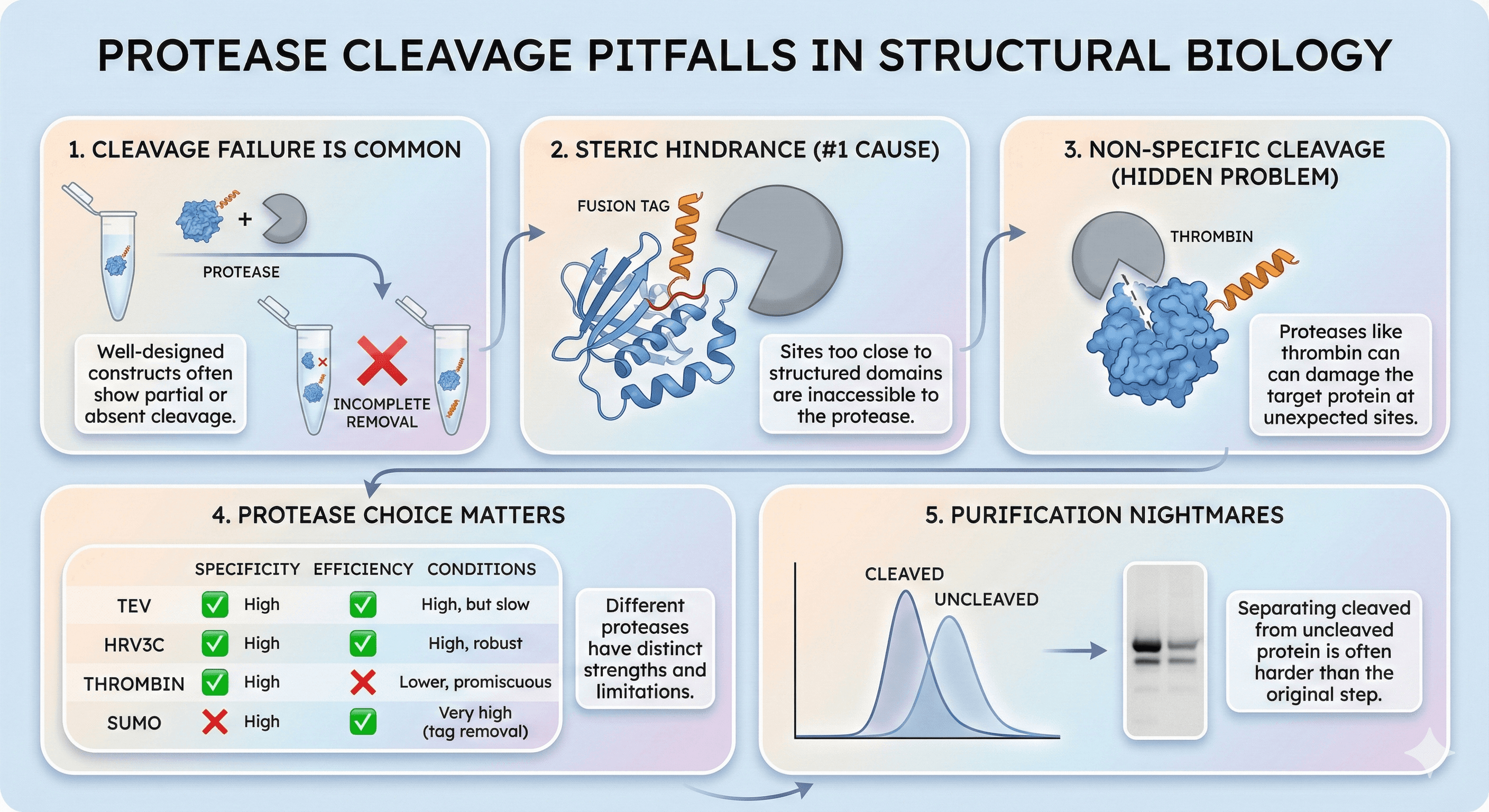
Protease cleavage failure is common: Even well-designed constructs frequently show incomplete or absent tag removal
Steric hindrance is the #1 cause: Cleavage sites too close to structured regions are inaccessible
Non-specific cleavage is a hidden problem: Proteases like thrombin can damage your protein at unexpected sites
Protease choice matters: TEV, HRV3C, thrombin, and SUMO protease have different strengths and limitations
Incomplete cleavage creates purification nightmares: Separating cleaved from uncleaved protein is often harder than the original purification
The Tag Removal Challenge
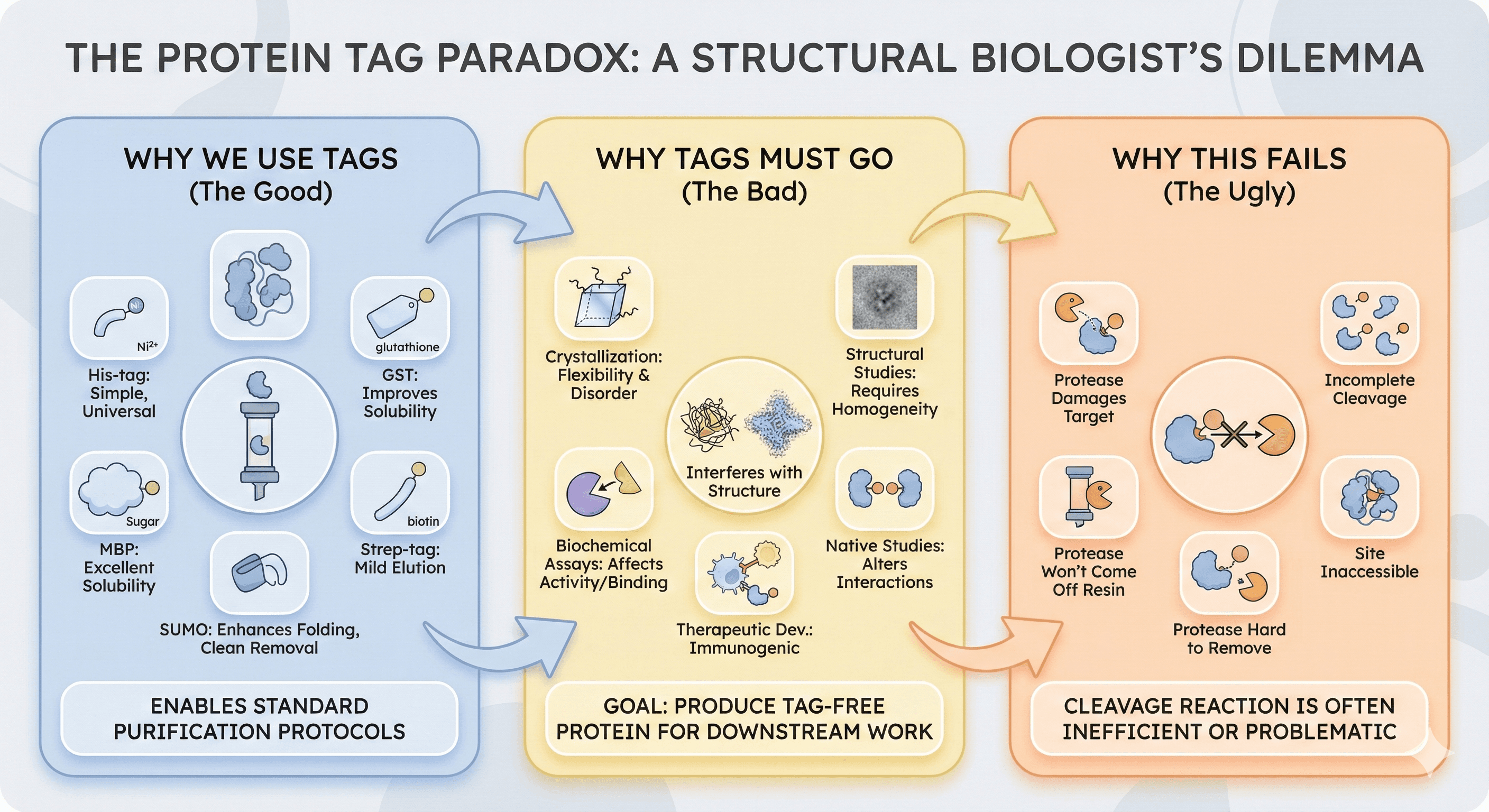
Why We Use Tags
Affinity tags revolutionized protein purification:
His-tag: Simple, universal, small
GST: Improves solubility, large but easy to remove
MBP: Excellent solubility enhancement
SUMO: Enhances folding, clean removal
Strep-tag: Mild elution conditions
Without tags, purification requires protein-specific method development. With tags, almost any protein can be purified with standard protocols.
Why Tags Must Go
Tags interfere with:
Crystallization: Tags introduce flexibility and disorder
Structural studies: Cryo-EM and X-ray require homogeneous samples
Biochemical assays: Tags can affect activity, binding, or localization
Therapeutic development: Non-native sequences are immunogenic
Native studies: Tags may alter oligomeric state or interactions
The goal: Capture the advantages of tagged expression, then produce tag-free protein for downstream work.
Why This Fails
The cleavage reaction seems simple:
In practice:
Protease can't access the cleavage site
Protease damages the target protein
Cleavage is incomplete
Protease won't come off the resin
Protease is as hard to remove as the tag
The Five Failure Modes
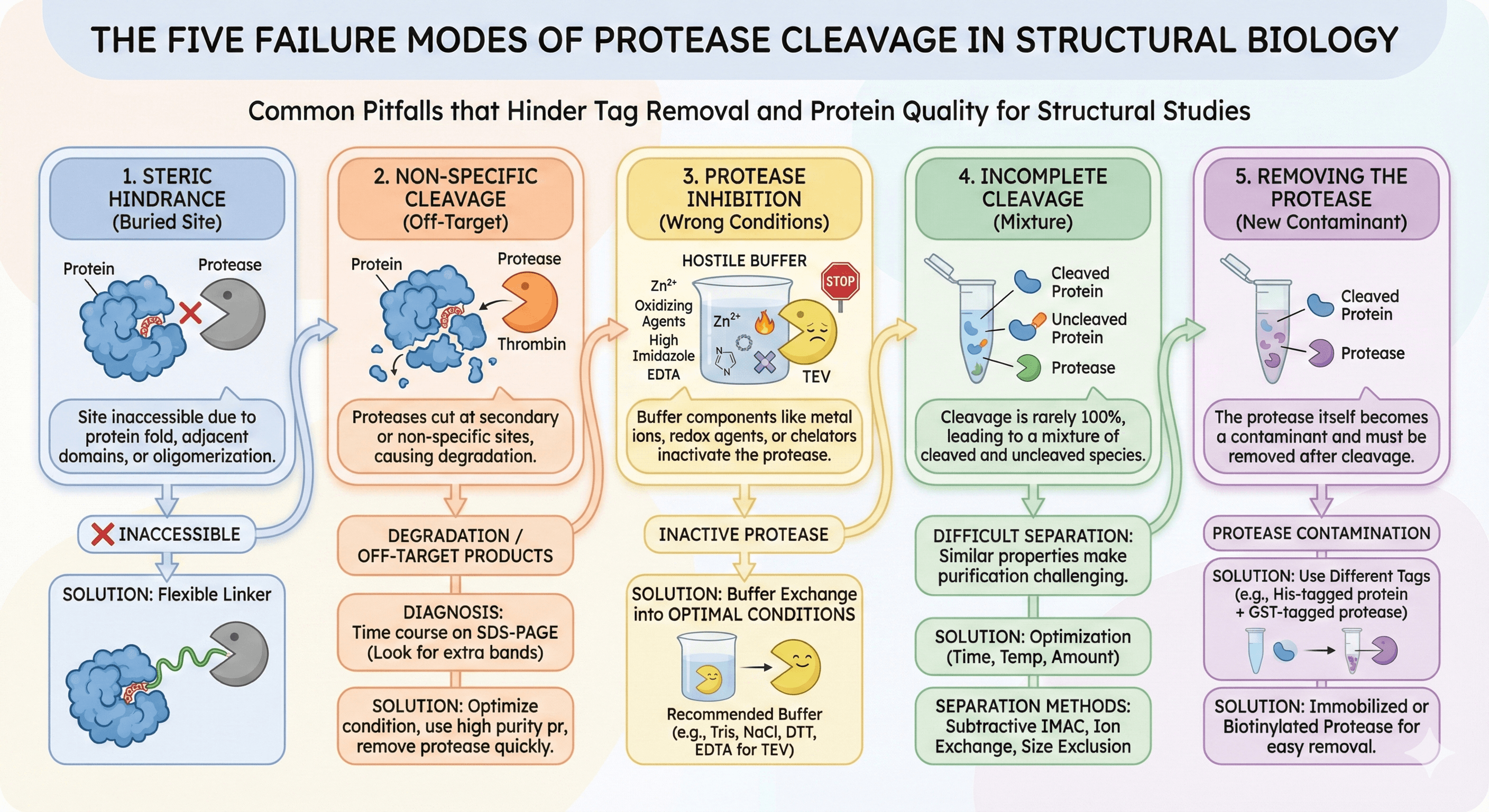
Failure 1: Steric Hindrance (The Cleavage Site Is Buried)
The problem: The protease needs to access and bind to the cleavage site. If the site is:
Too close to folded structure
Partially buried by the protein surface
Occluded by adjacent domains
Blocked by oligomerization
...the protease physically cannot reach it.
The evidence: Research on affinity tag removal has shown that "the inability of a protease to cleave a fusion protein may be caused by steric hindrance, which can be of several types. For example, the cleavage site may be too close to ordered structure in the target protein."
The problem is especially severe for oligomeric proteins. Even tags as small as polyhistidine can create steric problems when present on multiple subunits of a multimer.
The solution: Insert a flexible linker between the cleavage site and your protein. Studies have demonstrated that adding five glycine residues between the TEV protease recognition site and the target protein dramatically improved cleavage efficiency.
The design principle:
Failure 2: Non-Specific Cleavage (The Protease Damages Your Protein)
The problem: Proteases aren't perfectly specific. They have:
Primary site: The designed cleavage site
Secondary sites: Weaker recognition sequences elsewhere in your protein
Non-specific activity: Especially at high concentrations or long incubations
Thrombin is the worst offender: Research on thrombin-induced degradation found that "while thrombin and factor Xa are quite specific for cleavage at the inserted cleavage site, proteolysis can frequently occur at other site(s) in the protein of interest."
The problem is compounded by commercial thrombin preparations containing secondary protease activity from contaminating proteases.
TEV isn't immune: Recent studies have revealed unexpected TEV protease cleavage of recombinant human proteins at non-canonical sites. Using broader sequence specificity rules, researchers identified 456 human proteins that could be substrates for unwanted TEV cleavage.
The diagnostic approach:
Run time course: Remove samples at 1h, 2h, 4h, overnight
Analyze by SDS-PAGE
Look for: Extra bands (degradation products), decreasing band intensity (target destruction)
The solutions:
Problem | Solution |
|---|---|
Thrombin secondary cleavage | Use heparin-Sepharose to remove thrombin immediately after cleavage |
TEV off-target sites | Check sequence for canonical and non-canonical sites before design |
General over-digestion | Optimize protease:substrate ratio; less is often more |
Contaminating proteases | Use high-purity protease sources |
Failure 3: Protease Inhibition (Your Buffer Kills the Enzyme)
The problem: Proteases have specific requirements and sensitivities:
Protease | Inhibited By | Requires |
|---|---|---|
TEV | Zn²⁺ >5 mM, iodoacetamide, oxidizing conditions | DTT (1 mM), EDTA (0.5 mM) |
HRV3C | Oxidizing conditions | Reducing conditions |
Thrombin | EDTA, DTT, high imidazole | Ca²⁺, appropriate pH |
Factor Xa | EDTA, high salt | Ca²⁺ |
SUMO protease | Varies by source | Depends on specific enzyme |
Common mistakes:
IMAC elution buffer: High imidazole inhibits thrombin
Reducing agents: DTT reduces thrombin activity
Metal chelators: EDTA kills metalloproteases
Wrong pH: All proteases have pH optima
The solution:
Always buffer exchange into protease-compatible conditions before cleavage. The elution buffer from your affinity step is rarely optimal for proteolysis.
50 mM Tris pH 8.0
150 mM NaCl
1 mM DTT
0.5 mM EDTA
Temperature: 4°C or room temperature
Ratio: 1:100 (TEV:substrate by OD280)
Failure 4: Incomplete Cleavage (You Get a Mixture)
The problem: Even when cleavage works, it's rarely 100%. You end up with:
60-90% cleaved protein
10-40% uncleaved protein
Same affinity tag on both
Nearly identical size
Why this is a nightmare: If cleavage is 80% complete:
Re-running Ni-NTA removes 20% of your protein (the uncleaved fraction)
But 80% of your cleaved protein also binds (weakly) due to native histidines
You lose most of your cleaved protein trying to remove the uncleaved fraction
The numbers: Studies report that His-tag cleavage is often "far from 100% efficient," with many proteins showing substantial uncleaved fractions even after optimization.
Solutions for improving cleavage:
More protease: But watch for non-specific cleavage
Longer incubation: But protein may aggregate or degrade
Higher temperature: TEV is maximally active at 34°C, but your protein may not survive
Fresh protease: Old protease loses activity
Solutions for separating cleaved/uncleaved:
Method | Principle | Challenge |
|---|---|---|
Subtractive IMAC | Uncleaved binds, cleaved flows through | Cleaved often has weak binding too |
Ion exchange | Charge difference | Small tag = small charge difference |
Size exclusion | Size difference | Tag often too small to resolve |
Reverse IMAC | His-tag protease binds, both proteins flow through | Need His-tagged protease |
Failure 5: Removing the Protease (Now You Have a New Contaminant)
The problem: After cleavage, your sample contains:
Cleaved target protein
Uncleaved target protein
Free tag
Protease
If the protease has the same tag as your protein (common with His-tagged TEV), standard subtractive purification won't work—both bind or both flow through together.
The on-column cleavage trap: On-column cleavage (adding protease while protein is on the affinity resin) seems elegant but often fails because the tag-removal protease binds the resin, sterically limiting its proteolytic activity.
Solutions:
Use differently tagged protease:
His-tagged protein + GST-tagged TEV
Then: Ni-NTA (removes uncleaved protein) → Glutathione (removes TEV)
Use untagged protease:
Commercially available
Requires additional purification step to separate by size or charge
Use immobilized protease:
Protease covalently attached to resin
Cleavage in batch, then filter
Protease stays on resin
Use biotinylated protease:
Cleave in solution
Remove protease with streptavidin beads
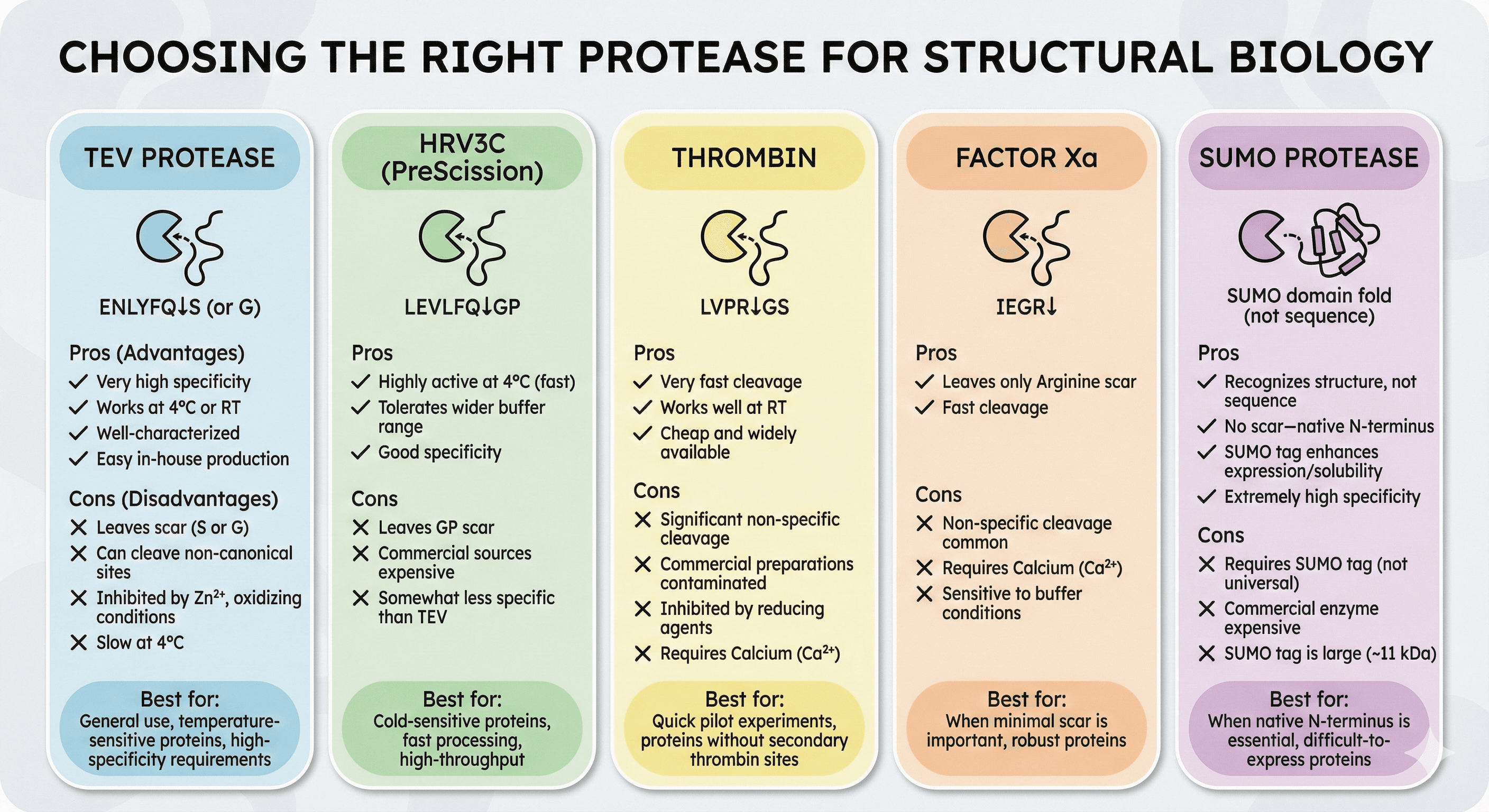
Choosing the Right Protease
TEV Protease
Recognition site: ENLYFQ↓S (or G)
Advantages:
Very high specificity
Works at 4°C (slowly) or room temperature (faster)
Well-characterized
Easy to produce in-house
Disadvantages:
Leaves scar (serine or glycine on target N-terminus)
Can cleave non-canonical sites in some proteins
Inhibited by zinc, oxidizing conditions
Slow at 4°C (overnight typical)
Best for: General use, temperature-sensitive proteins, high-specificity requirements
HRV3C (PreScission) Protease
Recognition site: LEVLFQ↓GP
Advantages:
Highly active at 4°C (minutes to hours)
Tolerates wider range of buffer additives than TEV
Good specificity
Disadvantages:
Leaves GP scar on target
Commercial sources can be expensive
Somewhat less specific than TEV
Best for: Cold-sensitive proteins, fast processing, high-throughput
Thrombin
Recognition site: LVPR↓GS
Advantages:
Very fast cleavage
Works well at room temperature
Cheap and widely available
Disadvantages:
Commercial preparations often contaminated
Inhibited by reducing agents
Requires calcium
Best for: Quick pilot experiments, proteins without secondary thrombin sites
Factor Xa
Recognition site: IEGR↓
Advantages:
Leaves only arginine attached to target
Fast cleavage
Disadvantages:
Non-specific cleavage common
Requires calcium
Sensitive to buffer conditions
Best for: When minimal scar is important, robust proteins
SUMO Protease
Recognition site: SUMO domain fold (not sequence)
Advantages:
Recognizes structure, not sequence
No scar—native N-terminus
SUMO tag enhances expression and solubility
Extremely high specificity
Disadvantages:
Requires SUMO tag (not universal)
Commercial enzyme expensive
SUMO tag is large (~11 kDa)
Best for: When native N-terminus is essential, difficult-to-express proteins
The Troubleshooting Decision Tree
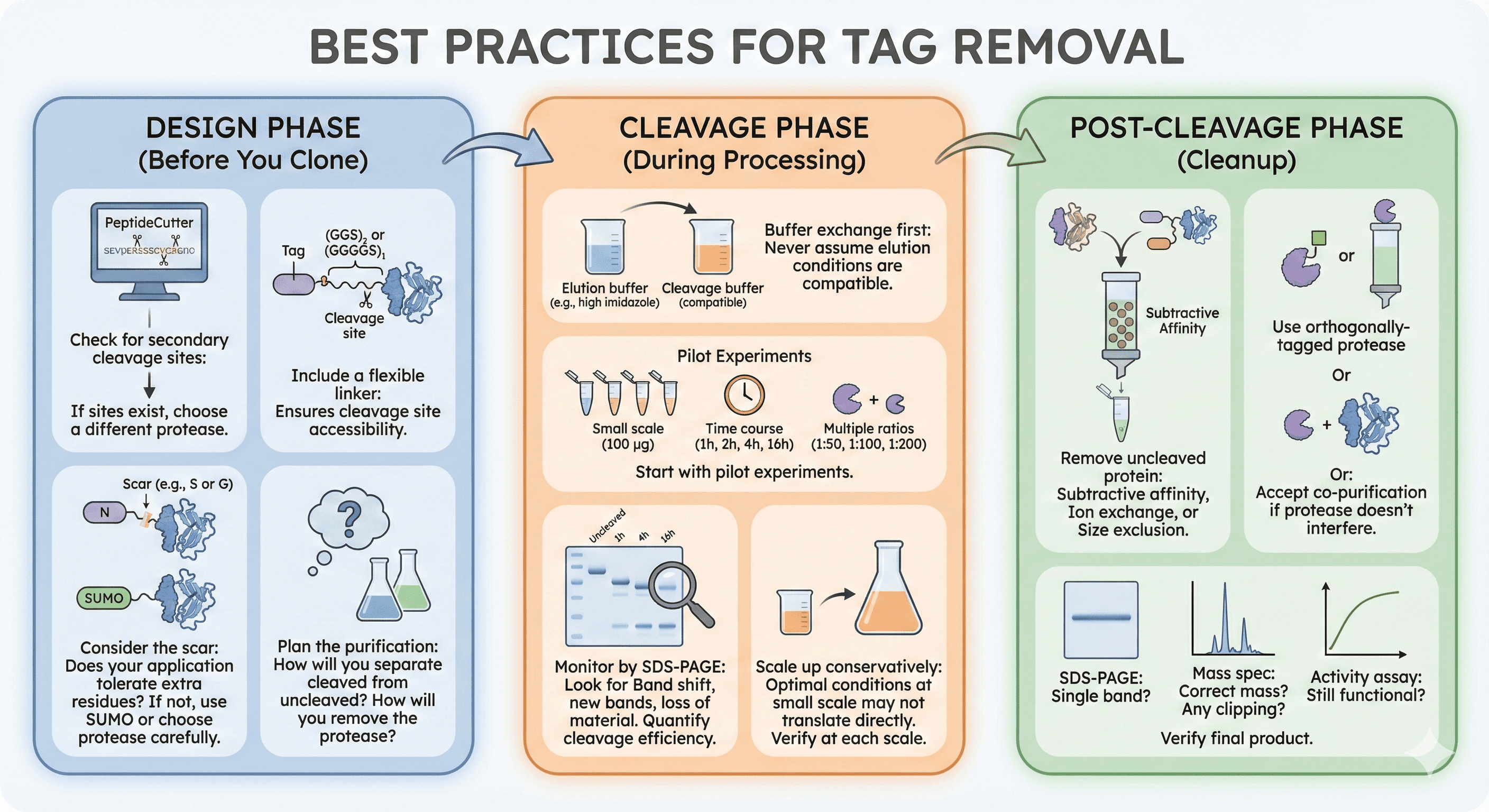
Best Practices for Tag Removal
Design Phase (Before You Clone)
Check for secondary cleavage sites:
Use PeptideCutter or similar tools
If sites exist, choose a different protease
Include a flexible linker:
(GGS)₂ or (GGGGS)₁ between tag and protein
Ensures cleavage site accessibility
Consider the scar:
Does your application tolerate extra residues at the N-terminus?
If not, use SUMO or choose protease carefully
Plan the purification:
How will you separate cleaved from uncleaved?
How will you remove the protease?
Cleavage Phase (During Processing)
Buffer exchange first:
Move from elution buffer to cleavage buffer
Never assume elution conditions are compatible
Start with pilot experiments:
Small scale (100 µg)
Time course (1h, 2h, 4h, 16h)
Multiple protease ratios (1:50, 1:100, 1:200)
Monitor by SDS-PAGE:
Look for: Band shift, new bands, loss of material
Quantify cleavage efficiency
Scale up conservatively:
Optimal conditions at small scale may not translate directly
Verify at each scale
Post-Cleavage Phase (Cleanup)
Remove uncleaved protein:
Subtractive affinity (if cleaved protein doesn't bind)
Ion exchange (if charge difference sufficient)
Size exclusion (if size difference sufficient)
Remove protease:
Use orthogonally-tagged protease
Or: Accept co-purification if protease doesn't interfere
Verify final product:
SDS-PAGE: Single band?
Mass spec: Correct mass? Any clipping?
Activity assay: Still functional?
Case Studies
Case 1: TEV Won't Cut
Protein: 45 kDa enzyme Construct: His6-TEV site-Target Problem: No cleavage after 24 hours with excess TEV
Diagnosis:
Crystal structure of homolog showed N-terminus buried in a groove
TEV site was inaccessible to protease
Solution:
Redesigned: His6-(GGGGS)₂-TEV site-(GGGGS)₂-Target
Cleavage now complete in 4 hours
Case 2: Thrombin Destroys the Protein
Protein: 30 kDa signaling domain Construct: His6-Thrombin site-Target Problem: Multiple bands after cleavage; activity lost
Diagnosis:
Target protein contained internal thrombin-like site
Both sites cleaved, fragmenting the protein
Solution:
Switched to TEV protease
No internal TEV sites present
Clean cleavage, full activity retained
Case 3: Can't Separate Cleaved from Uncleaved
Protein: 25 kDa structural protein Construct: His6-TEV site-Target Cleavage: 75% complete (couldn't improve further)
Problem: Cleaved protein has native His-rich region, binds Ni-NTA
Diagnosis:
Subtractive purification impossible
Both species bind column
Solution:
Switched to Strep-tag
StrepTactin binding is tag-specific
Cleaved protein flows through cleanly
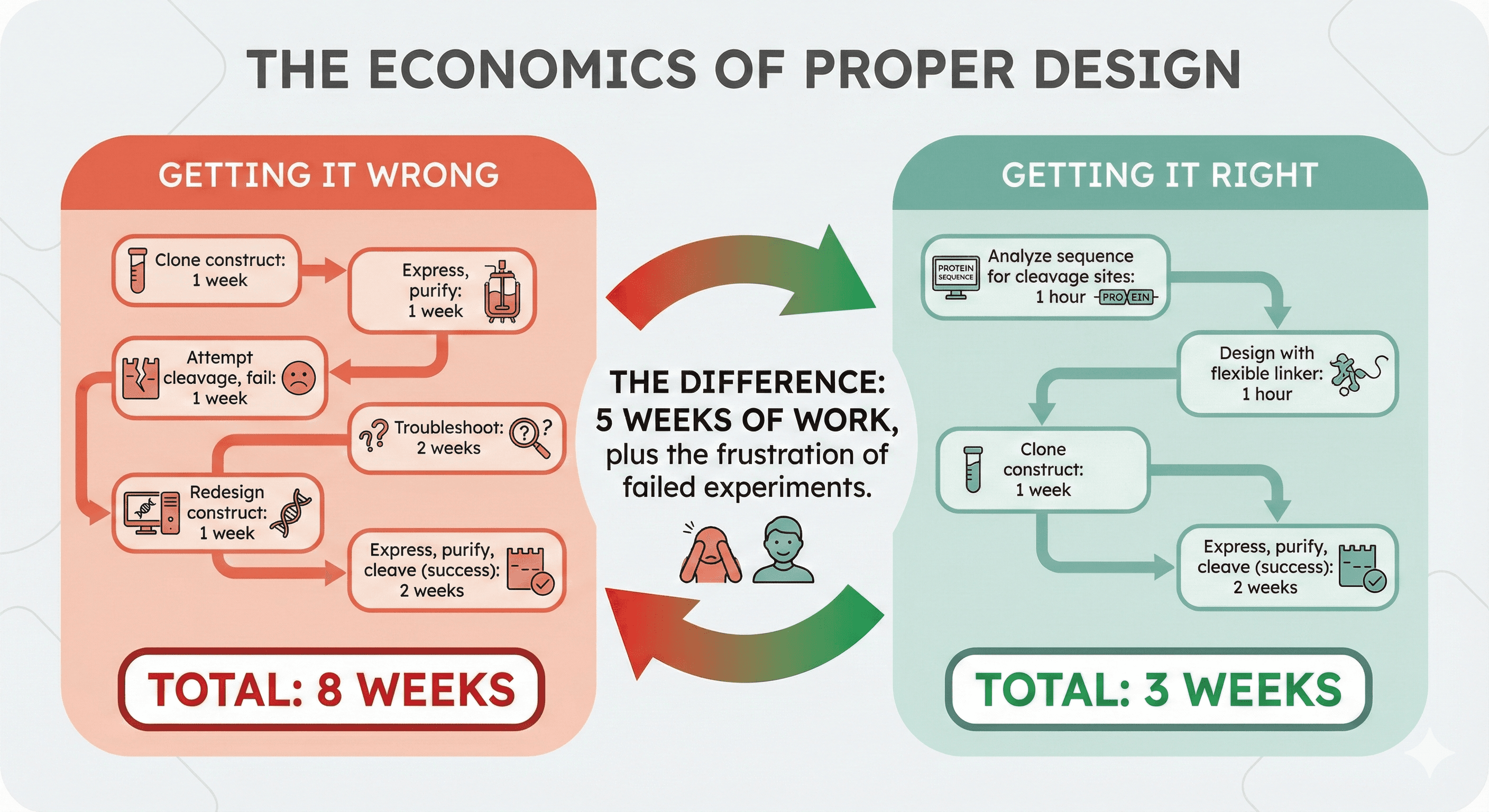
The Economics of Proper Design
Getting It Wrong
Clone construct: 1 week
Express, purify: 1 week
Attempt cleavage, fail: 1 week
Troubleshoot: 2 weeks
Redesign construct: 1 week
Express, purify, cleave (success): 2 weeks
Total: 8 weeks
Getting It Right
Analyze sequence for cleavage sites: 1 hour
Design with flexible linker: 1 hour
Clone construct: 1 week
Express, purify, cleave (success): 2 weeks
Total: 3 weeks
The difference: 5 weeks of work, plus the frustration of failed experiments.
The Bottom Line
Tag removal failure has predictable causes:
Cause | Prevention |
|---|---|
Steric hindrance | Design with flexible linkers |
Wrong buffer | Exchange before cleavage |
Non-specific cleavage | Check sequence, optimize conditions |
Incomplete cleavage | Pilot experiments, optimization |
Protease contamination | Plan removal strategy before starting |
The most common mistake is assuming cleavage will work without optimization. Every protein is different. What works for one construct may fail completely for another.
Design for success: check sequences, include linkers, plan your purification, and always run pilot experiments before committing to large-scale cleavage.
Construct Design Considerations
For researchers designing expression constructs, platforms like Orbion can help identify potential problems before cloning:
Secondary cleavage site detection: Identify sequences that match protease recognition patterns
Terminus accessibility analysis: Predict whether N- and C-termini are accessible or structured
PTM site mapping: Ensure cleavage sites don't overlap with essential modifications
Disorder prediction: Identify flexible regions suitable for tag placement
Good construct design prevents most tag removal problems—and good design starts with understanding your protein's structure and sequence features before you order the first oligonucleotide.
References
Waugh DS. (2011). An overview of enzymatic reagents for the removal of affinity tags. Protein Expression and Purification, 80(2):283-293. PMC3195948
Feehan RP & Bhattacharya S. (2024). Unexpected tobacco etch virus (TEV) protease cleavage of recombinant human proteins. Protein Expression and Purification, 220:106498. PMC11129917
Peti W & Page R. (2007). Strategies to maximize heterologous protein expression in Escherichia coli with minimal cost. Protein Expression and Purification, 51(1):1-10.
Ahuja S, et al. (2008). A method for the prevention of thrombin-induced degradation of recombinant proteins. Analytical Biochemistry, 382(1):67-69. PMC2614318
Kapust RB & Waugh DS. (2017). Removal of affinity tags with TEV protease. Methods in Molecular Biology, 1586:221-241. PMC7974378
Kronqvist N, et al. (2020). NT*-HRV3CP: An optimized construct of human rhinovirus 14 3C protease for high-yield expression and fast affinity-tag cleavage. Journal of Biotechnology, 323:109-117. ScienceDirect
Sigma-Aldrich. (2024). Tag removal proteases for recombinant protein purification. Technical Document. Link