Blog
Why High-Throughput Screening Hits Don't Reproduce
Feb 9, 2026
The primary screen identified 847 hits. Dose-response narrowed it to 127 confirmed actives. You cherry-picked the best 20 for follow-up. Six months later, you've tested them all in orthogonal assays. Exactly two show real activity against your target. The other 18 were artifacts, aggregators, or assay interference. A 90% false positive rate—and you're not alone.
This is the reproducibility crisis in high-throughput screening, and it's one of the most expensive problems in early drug discovery.
Key Takeaways
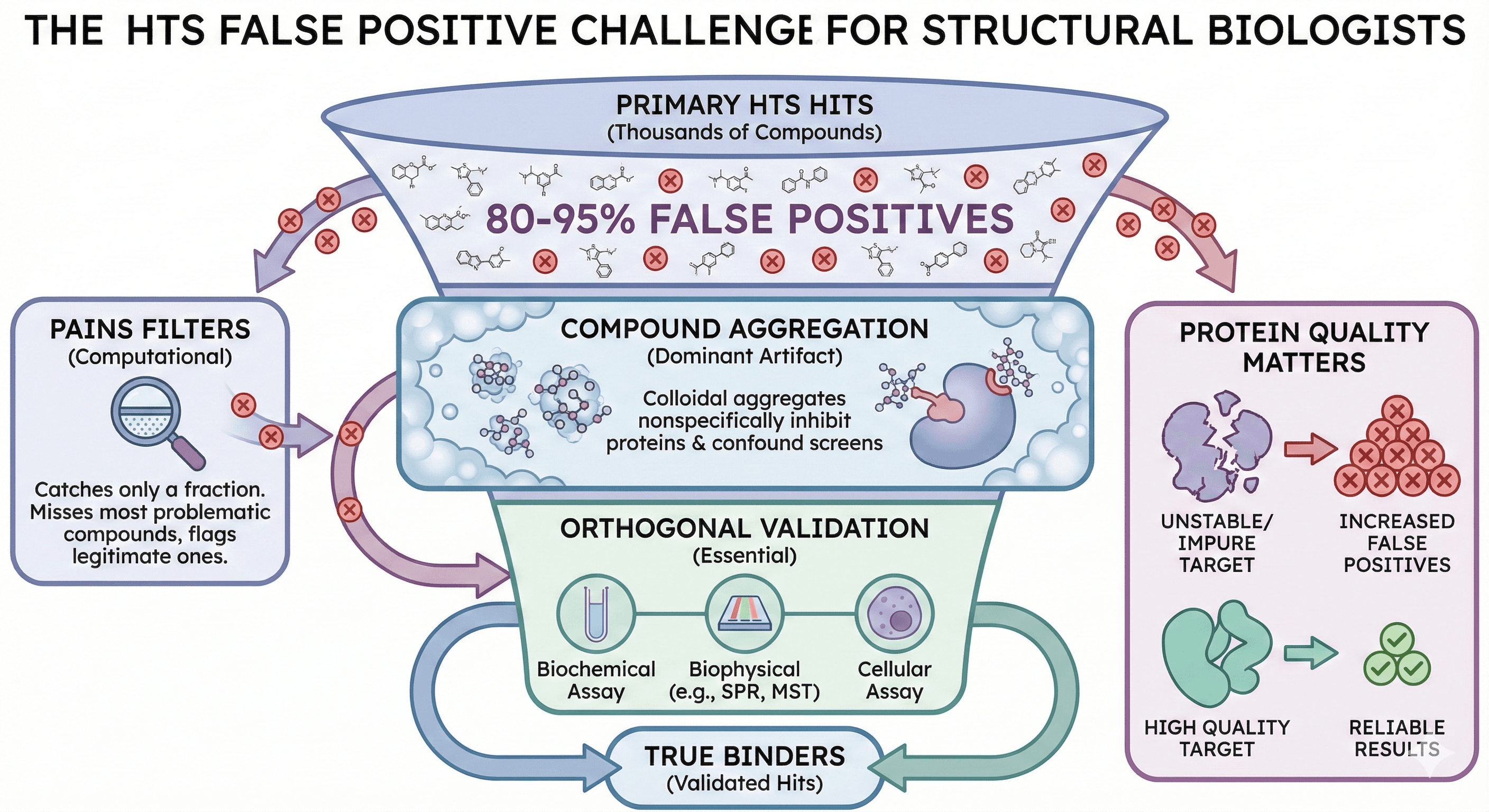
Most HTS hits are false positives: Industry estimates suggest 80-95% of primary hits fail to reproduce in orthogonal assays
Compound aggregation is the dominant artifact: Colloidal aggregates nonspecifically inhibit proteins and confound most biochemical screens
PAINS filters catch only a fraction: Computational filters miss most problematic compounds and flag many legitimate ones
Orthogonal validation is essential: No single assay type can distinguish true binders from artifacts
Protein quality matters: Unstable or impure target protein increases false positive rates dramatically
The False Positive Problem
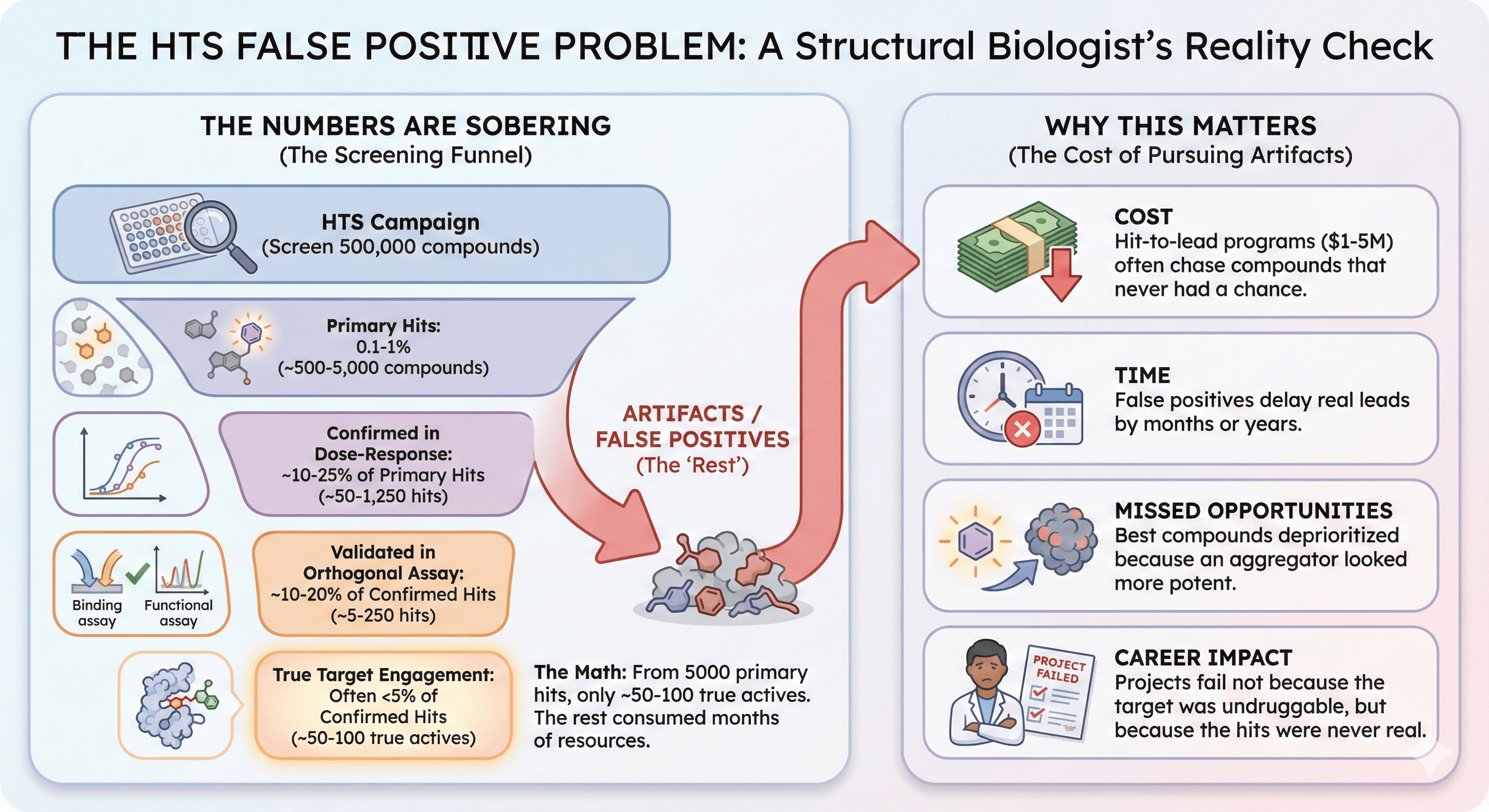
The Numbers Are Sobering
A typical HTS campaign:
Screen 500,000 compounds
Primary hits: 0.1-1% (~500-5000 compounds)
Confirmed in dose-response: ~10-25% of primary hits
Validated in orthogonal assay: ~10-20% of confirmed hits
True target engagement: Often <5% of confirmed hits
The math: From 5000 primary hits, you might get 50-100 true actives. The rest consumed months of chemistry resources pursuing artifacts.
Why This Matters
Cost: Hit-to-lead programs cost $1-5 million. Most are chasing compounds that never had a chance.
Time: False positives delay real leads by months or years.
Missed opportunities: The best compounds might have been deprioritized because an aggregator looked more potent.
Career impact: Projects fail not because the target was undruggable, but because the hits were never real.
The Five Sources of False Positives
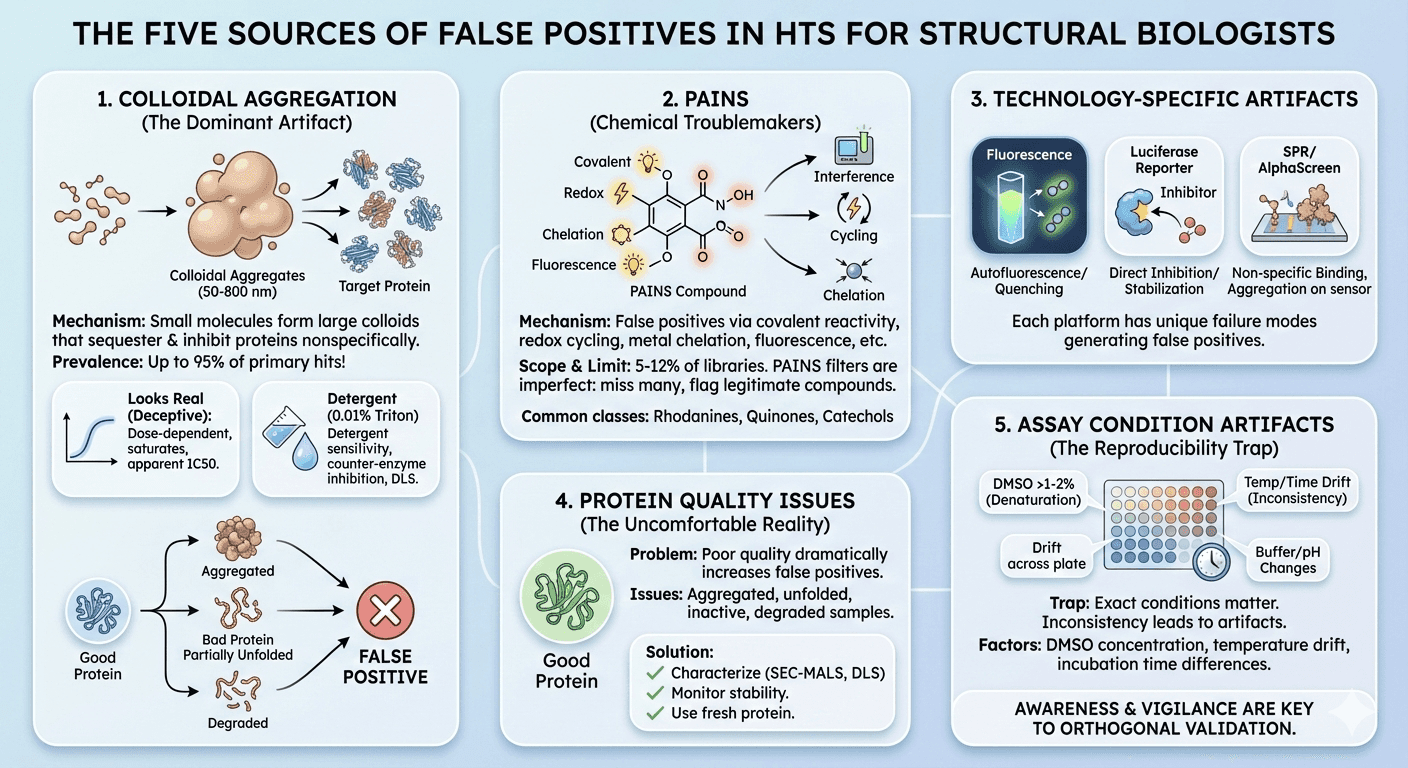
Source 1: Colloidal Aggregation
The mechanism: Many organic molecules—including drugs, clinical candidates, and especially early leads—spontaneously form colloids in aqueous buffer. These aggregates are densely packed particles ranging from 50 to over 800 nm in radius that sequester 10⁵ to 10⁶ protein molecules, leading to their partial denaturation and typically their inhibition.
The prevalence: Studies suggest that aggregation has been reported to occur for up to 95% of primary hits from HTS campaigns. This is the dominant source of false positives in biochemical screens.
The insidious nature: Aggregation-based inhibition looks real:
Dose-dependent (more compound = more aggregates = more inhibition)
Saturates at high concentration
Shows apparent IC50 values
Can be potent (sub-micromolar apparent IC50)
What doesn't distinguish it from true inhibitors:
Apparent potency
Hill slope (often ~1)
Time-dependence (if any)
What does distinguish it:
Detergent sensitivity: Add 0.01-0.1% Triton X-100 or Tween-80; aggregate-based inhibition disappears
Counter-enzyme susceptibility: Aggregators inhibit unrelated enzymes (e.g., AmpC β-lactamase as a counter-screen)
Dynamic light scattering (DLS): Particles visible above critical aggregation concentration
Even cell-based assays aren't immune: Recent research on COVID-19 drug repurposing showed that of 41 tested drugs, 17 behaved as classic aggregators, forming particles by DLS and inhibiting counter-screening enzymes. Colloidal aggregation contributed to false positives in cell-based antiviral screens.
Source 2: Pan-Assay Interference Compounds (PAINS)
What PAINS are: PAINS are chemical compounds that give false positive results in many different assay types due to:
Covalent reactivity with proteins
Redox cycling
Metal chelation
Fluorescence interference
Photoreactivity
Common PAINS classes:
Rhodanines
Quinones
Catechols
Isothiazolones
Hydroxyphenyl hydrazones
Curcuminoids
Enones
The scope: Studies estimate that a typical academic screening library contains roughly 5-12% PAINS. There are approximately 400 structural classes of PAINS.
The limitation of PAINS filters: Large-scale analysis found that "the same PAINS substructure was often found in consistently inactive and frequently active compounds, indicating that the structural context in which PAINS occur modulates their effects."
PAINS filters are problematic: they are "oversensitive and disproportionately flag compounds as interference compounds while failing to identify a majority of truly interfering compounds." In other words, they create both false positives and false negatives in compound triage.
Better approaches: Quantitative Structure-Interference Relationship (QSIR) models for specific interference mechanisms (thiol reactivity, redox activity, luciferase activity) show 58-78% external balanced accuracy—better than PAINS filters but still imperfect.
Source 3: Technology-Specific Artifacts
Different assay formats have different vulnerabilities:
Fluorescence-based assays:
Compound autofluorescence (false activation)
Fluorescence quenching (false inhibition)
Inner filter effects at high concentration
Compound fluorescence overlapping with probe
Luciferase reporter assays:
Luciferase inhibition (independent of pathway)
Luciferase stabilization
Compound effects on reporter expression
Redox-active compounds
AlphaScreen/HTRF:
Singlet oxygen scavengers (false inhibition)
Light absorbers
Compound aggregation
Metal chelation affecting donor/acceptor beads
SPR (Surface Plasmon Resonance):
Non-specific binding to chip surface
Aggregation on sensor
Refractive index changes
Compound precipitation in flow
Each technology has specific failure modes that generate false positives unique to that platform.
Source 4: Protein Quality Issues
The problem nobody wants to discuss: Poor protein quality dramatically increases false positive rates:
Protein Issue | How It Causes False Positives |
|---|---|
Aggregated protein | Binds compounds nonspecifically |
Partially unfolded | Exposes normally buried sites |
Inactive fraction | Shifts apparent IC50, flat SAR |
Degraded protein | Cleavage products may have altered binding |
Wrong oligomeric state | Different binding properties |
Missing cofactor | Dead enzyme, artifacts dominate |
The uncomfortable reality: Production of recombinant proteins is often hampered by instability and propensity to aggregate. Protein samples of poor quality are associated with reduced reproducibility.
When your target protein is 30% aggregated, 20% inactive, and slowly degrading during the screen—your data is compromised before a single compound is tested.
What to do:
Characterize protein quality before screening (SEC-MALS, DLS, activity assay)
Monitor stability over screening duration
Use fresh protein batches
Include positive control compounds on every plate
Track Z' factor for quality control
Source 5: Assay Condition Artifacts
The reproducibility trap: The exact conditions of screening matter enormously:
DMSO concentration: Above 1-2%, protein starts denaturing
Buffer composition: pH, ionic strength, divalent cations all affect binding
Temperature: Room temperature assays drift
Incubation time: Equilibrium may not be reached
Protein concentration: Below Kd, binding is hard to detect
Order of addition: Compounds added to protein vs. protein added to compounds
The drift problem: HTS assay validation guidelines emphasize that "plates should not exhibit material edge or drift effects." But in practice, a 384-well plate takes time to fill, and conditions change:
First wells incubate longer than last wells
Reagents in reservoirs warm up
DMSO evaporates, changing final concentration
Enzyme activity decays during plate setup
The Validation Cascade
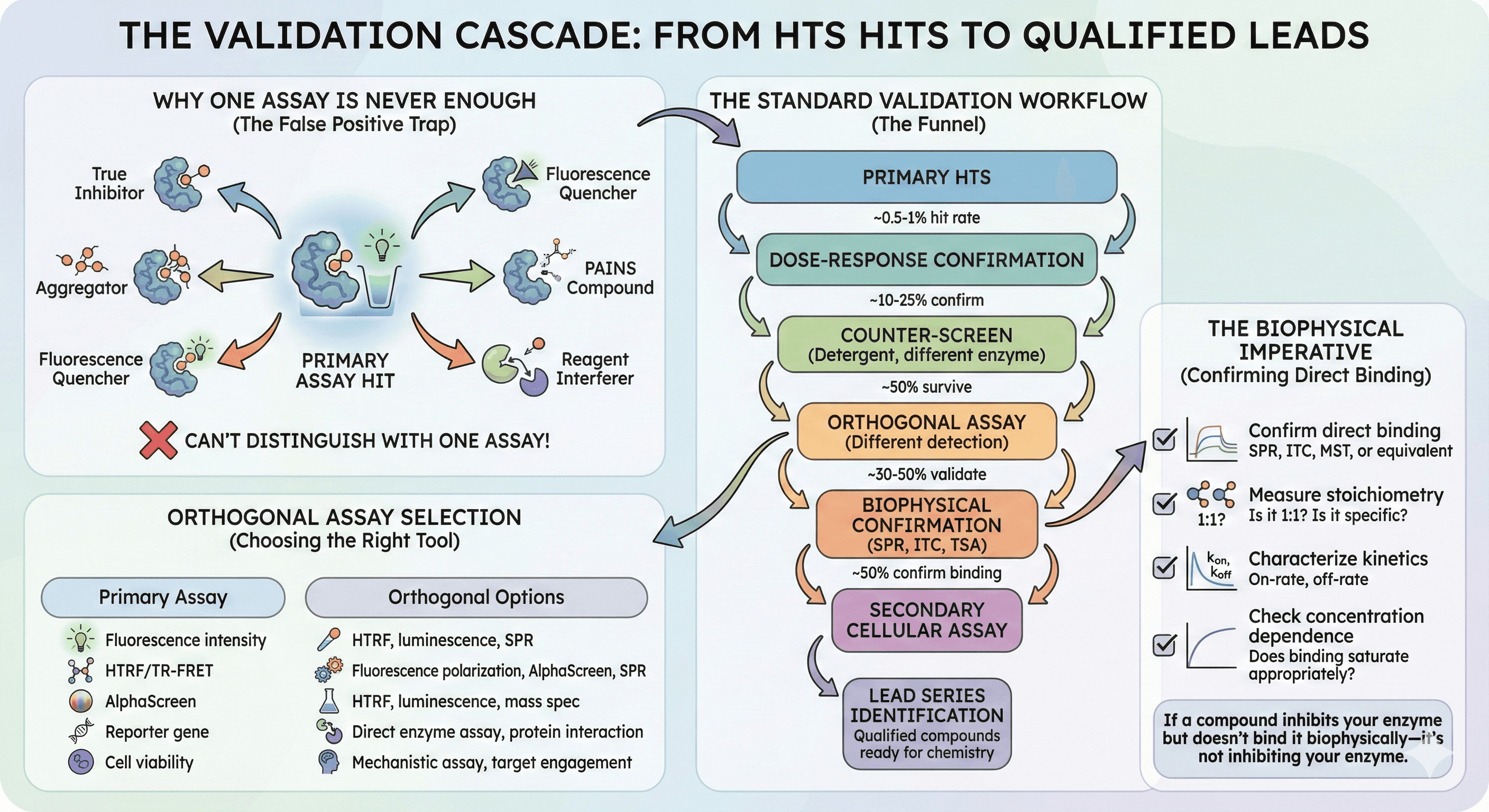
Why One Assay Is Never Enough
A compound that inhibits your target in a fluorescence assay might be:
A true inhibitor
An aggregator
A fluorescence quencher
A PAINS compound that reacts with your target (but also everything else)
A compound that affects assay reagents
You can't distinguish these with a single assay format.
The Standard Validation Workflow
Each step reduces compound numbers by 50-80%.
Orthogonal Assay Selection
The second assay should:
Use a different detection technology
Measure the same biochemical event
Be insensitive to the artifacts of the first assay
Primary Assay | Orthogonal Options |
|---|---|
Fluorescence intensity | HTRF, luminescence, SPR |
HTRF/TR-FRET | Fluorescence polarization, AlphaScreen, SPR |
AlphaScreen | HTRF, luminescence, mass spec |
Reporter gene | Direct enzyme assay, protein interaction |
Cell viability | Mechanistic assay, target engagement |
The Biophysical Imperative
For any hit going into chemistry:
Confirm direct binding: SPR, ITC, MST, or equivalent
Measure stoichiometry: Is it 1:1? Is it specific?
Characterize kinetics: On-rate, off-rate
Check concentration dependence: Does binding saturate appropriately?
If a compound inhibits your enzyme but doesn't bind it biophysically—it's not inhibiting your enzyme.
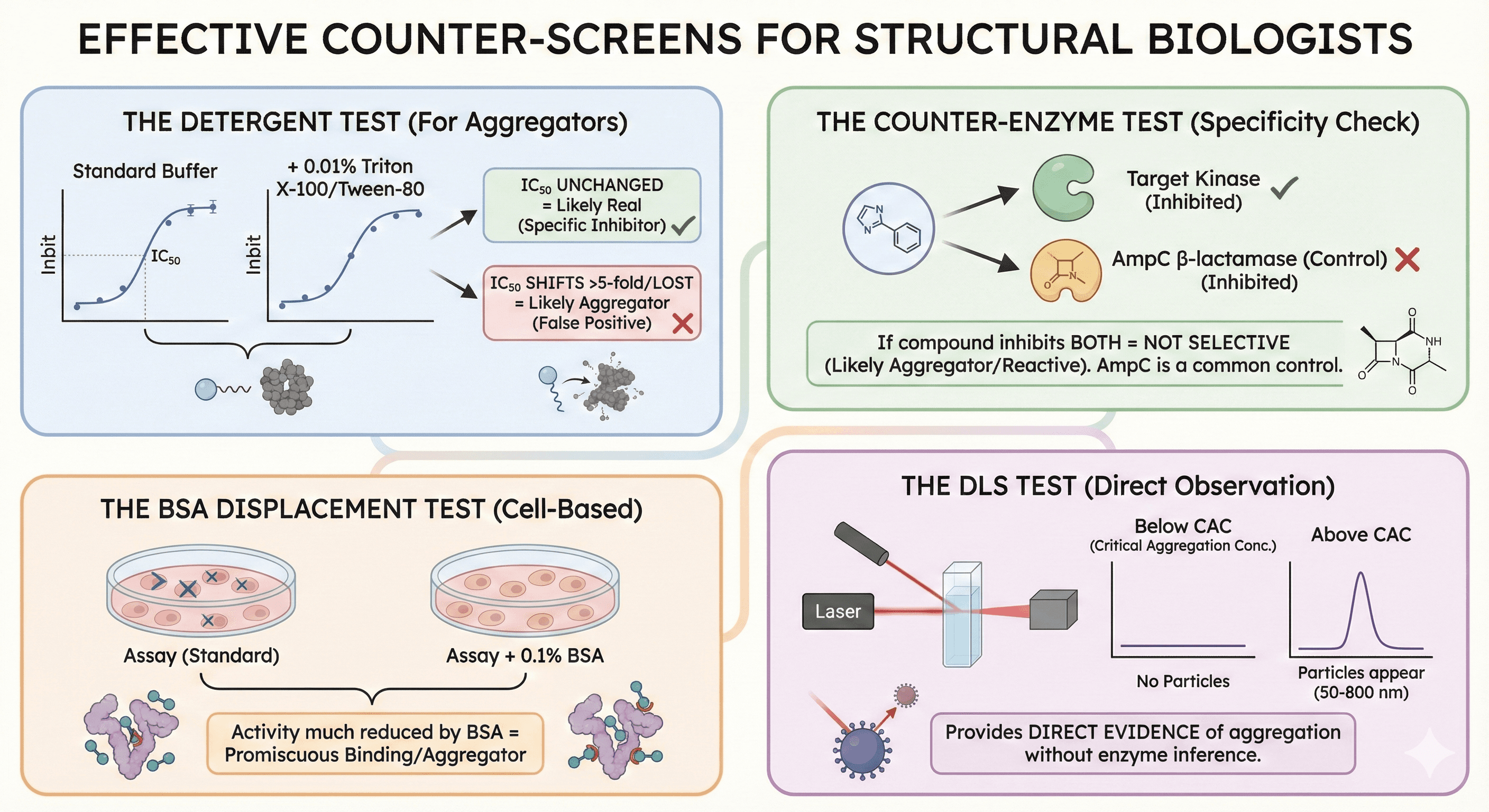
Counter-Screens That Work
The Detergent Test
Simple and effective for aggregators:
Run dose-response in standard buffer
Run dose-response with 0.01% Triton X-100 or Tween-80
Compare IC50 values
Interpretation:
IC50 unchanged: Likely real
IC50 shifts >5-fold or activity lost: Likely aggregator
Research has shown that this counter-screen reliably detects aggregate-based inhibition because non-ionic detergent disrupts the colloidal aggregates.
The Counter-Enzyme Test
Use an unrelated enzyme as a control: If your compound inhibits both your target kinase AND AmpC β-lactamase—it's not selective. It's probably an aggregator or reactive compound.
AmpC β-lactamase is commonly used because it's extensively characterized for aggregate-based inhibition.
The BSA Displacement Test
For cell-based assays: Run assay with and without 0.1% BSA (bovine serum albumin). Compounds whose activity is much reduced by BSA addition may be aggregators or have promiscuous protein binding.
The DLS Test
Direct observation of aggregation: Measure particle size by dynamic light scattering:
Below critical aggregation concentration: No particles
Above CAC: Particles appear (50-800 nm)
DLS provides direct evidence of aggregation without inference from enzyme inhibition.
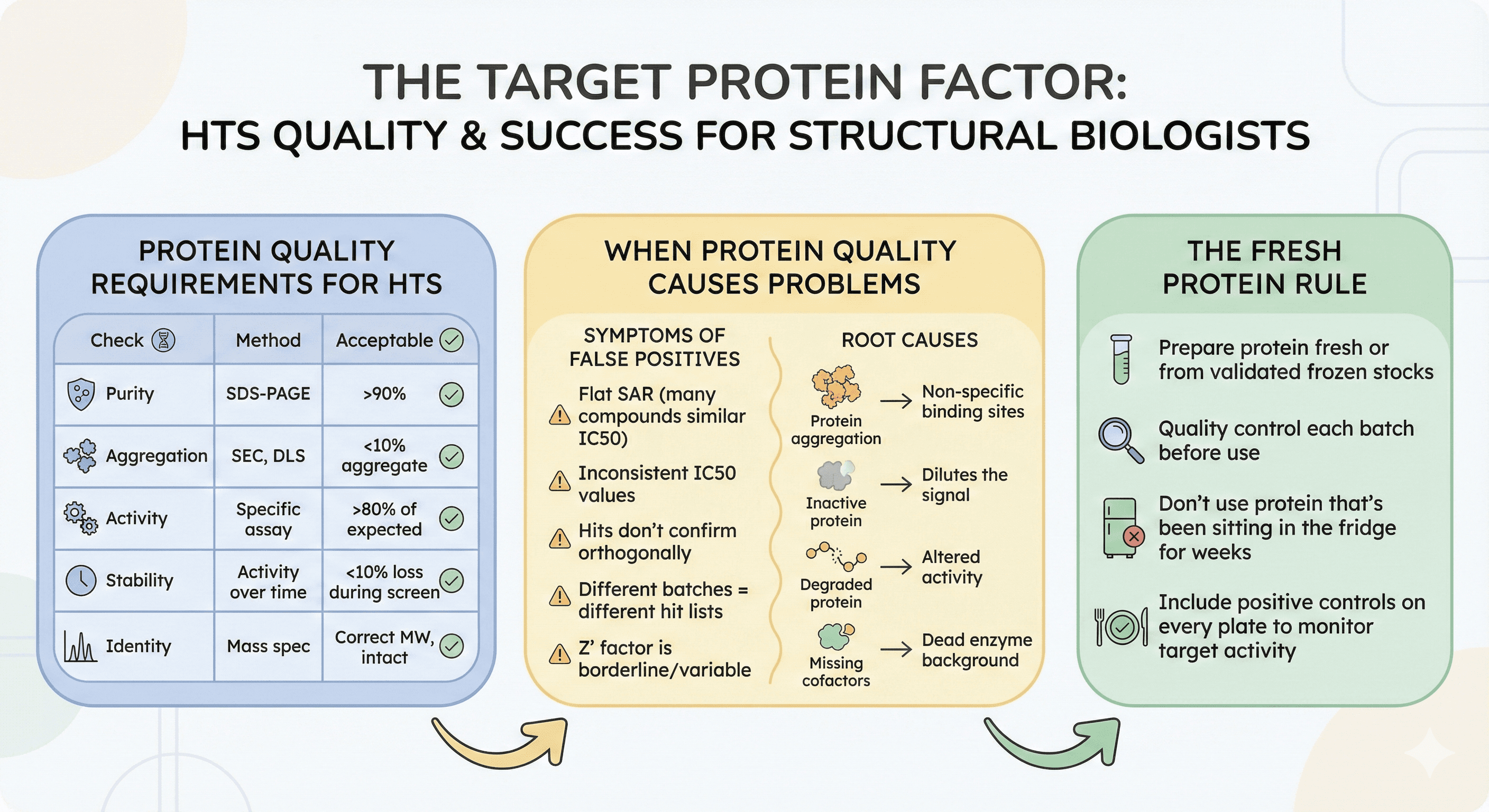
The Target Protein Factor
Protein Quality Requirements for HTS
Before screening, verify:
Check | Method | Acceptable |
|---|---|---|
Purity | SDS-PAGE | >90% |
Aggregation | SEC, DLS | <10% aggregate |
Activity | Specific assay | >80% of expected |
Stability | Activity over time | <10% loss during screen |
Identity | Mass spec | Correct MW, intact |
When Protein Quality Causes Problems
Symptoms of protein-derived false positives:
Flat SAR (many compounds have similar IC50)
IC50 values inconsistent with biochemical understanding
Hits don't confirm in orthogonal assays
Different protein batches give different hit lists
Z' factor is borderline or variable
Root causes:
Protein aggregation creates non-specific binding sites
Inactive protein dilutes the signal
Degraded protein has altered activity
Missing cofactors create dead enzyme background
The Fresh Protein Rule
For critical screens:
Prepare protein fresh or from validated frozen stocks
Quality control each batch before use
Don't use protein that's been sitting in the fridge for weeks
Include positive controls on every plate to monitor target activity
Case Studies in False Positives
Case 1: The Aggregator Epidemic
Screen: Kinase inhibitor HTS, 300,000 compounds Primary hits: 2,400 compounds (0.8%) Dose-response confirmed: 480 compounds (20%) After detergent counter-screen: 96 compounds (20% of confirmed) After SPR: 28 compounds confirmed binding (6% of original confirmed hits)
Lesson: 80% of confirmed hits were aggregators, eliminated by a simple detergent test.
Case 2: The Luciferase Artifact
Screen: Reporter gene assay for pathway activation Primary hits: 150 activators Counter-screen (constitutive luciferase): 120 also activated control reporter True pathway activators: 30 compounds (20% of hits)
Lesson: 80% of "activators" were stabilizing luciferase, not activating the pathway.
Case 3: The Fluorescent Compound
Screen: Fluorescence polarization for protein-protein interaction inhibitor Top hit: Apparent IC50 = 100 nM, beautiful dose-response Orthogonal SPR: No binding detected Investigation: Compound was fluorescent at assay wavelength, creating artifact
Lesson: The most potent hit was completely artifactual.
Case 4: The Protein Quality Problem
Screen: Enzyme inhibitor, fluorescence-based First campaign (old protein): 450 hits, 5% confirmed in orthogonal Second campaign (fresh protein): 180 hits, 35% confirmed in orthogonal
Lesson: Degraded protein had 2.5× more hits, but 7× fewer real ones.
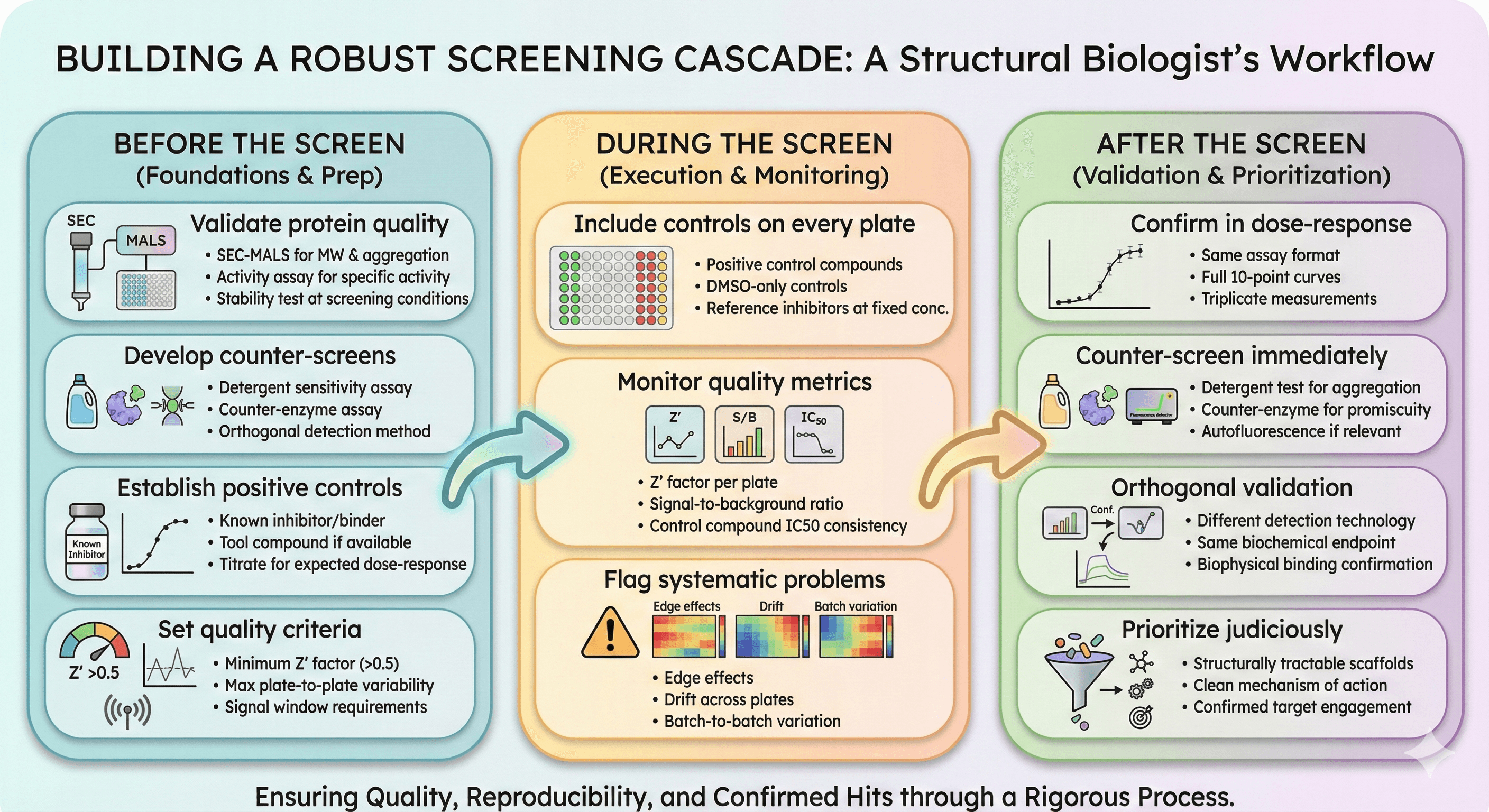
Building a Robust Screening Cascade
Before the Screen
Validate protein quality
SEC-MALS for MW and aggregation
Activity assay for specific activity
Stability test at screening conditions
Develop counter-screens
Detergent sensitivity assay
Counter-enzyme assay
Orthogonal detection method
Establish positive controls
Known inhibitor/binder
Tool compound if available
Titrate to establish expected dose-response
Set quality criteria
Minimum Z' factor (typically >0.5)
Maximum plate-to-plate variability
Signal window requirements
During the Screen
Include controls on every plate
Positive control compounds
DMSO-only controls
Reference inhibitors at fixed concentration
Monitor quality metrics
Z' factor per plate
Signal-to-background ratio
Control compound IC50 consistency
Flag systematic problems
Edge effects
Drift across plates
Batch-to-batch variation
After the Screen
Confirm in dose-response
Same assay format
Full 10-point curves
Triplicate measurements
Counter-screen immediately
Detergent test for aggregation
Counter-enzyme for promiscuity
Autofluorescence if relevant
Orthogonal validation
Different detection technology
Same biochemical endpoint
Biophysical binding confirmation
Prioritize judiciously
Structurally tractable scaffolds
Clean mechanism of action
Confirmed target engagement
The Bottom Line
False positives are not failures of the screen—they're expected outcomes that require systematic elimination. The question isn't whether you'll have false positives (you will); it's whether you'll identify them before investing months in medicinal chemistry.
Prevention Strategy | Implementation |
|---|---|
Aggregation | Detergent counter-screen, DLS |
PAINS | Structural filters (with caution), QSIR models |
Technology artifacts | Orthogonal assay validation |
Protein quality | QC before screening, batch-to-batch controls |
Assay artifacts | Robust validation, positive controls |
The key insight: A hit is not a hit until it's validated by orthogonal methods. Primary screening identifies candidates for investigation—not compounds ready for optimization.
Target Validation for Screening Campaigns
For researchers planning HTS campaigns, understanding your target protein is the first line of defense against false positives. Platforms like Orbion can help with:
Aggregation propensity prediction: Identify whether your target is likely to aggregate, affecting assay performance
Binding site analysis: Understand the druggable pockets you're screening against
Protein stability assessment: Predict regions that may cause instability during screening
PTM requirements: Identify modifications that might be essential for proper target behavior
Better target understanding leads to better assay design, which leads to fewer false positives—saving months of wasted effort chasing artifacts.
References
Shoichet BK. (2006). Screening in a spirit haunted world. Drug Discovery Today, 11(13-14):607-615. PMC4646424
Owen SC, et al. (2012). Colloidal aggregation causes inhibition of G protein-coupled receptors. Journal of Medicinal Chemistry, 55(16):7203-7211. PMC3613083
Feng BY & Bhatt R. (2006). A detergent-based assay for the detection of promiscuous inhibitors. Nature Protocols, 1(2):550-553. PMC1544377
Tran H, et al. (2023). Colloidal aggregation confounds cell-based Covid-19 antiviral screens. ACS Chemical Biology, 18(10):2332-2342. PMC10634915
Baell JB & Holloway GA. (2010). New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. Journal of Medicinal Chemistry, 53(7):2719-2740.
Jasial S, et al. (2017). How frequently are pan-assay interference compounds active? Large-scale analysis of screening data reveals diverse activity profiles, low global hit frequency, and many consistently inactive compounds. Journal of Medicinal Chemistry, 60(9):3879-3886. Link
Baell JB & Walters MA. (2014). Chemical con artists foil drug discovery. Nature, 513:481-483. Link
Schorpp K, et al. (2020). High-throughput screening to predict chemical-assay interference. Scientific Reports, 10:3839. Link
Iversen PW, et al. (2012). HTS assay validation. In: Assay Guidance Manual. Eli Lilly & Company and NIH NCATS. NCBI Bookshelf
Raynal B, et al. (2022). Measuring protein aggregation and stability using high-throughput biophysical approaches. Frontiers in Molecular Biosciences, 9:890862. Link

