Blog
Your Protein Is a Dimer in Solution But a Monomer in the Crystal
Feb 6, 2026
The SEC column says your protein is 85 kDa—clearly a dimer of your 42 kDa subunit. You solve the crystal structure. It's a monomer. The crystallographers say the oligomeric state is artifactual. The biochemists say the crystal is wrong. Both are confident. Both might be right—or both might be wrong.
Determining the true oligomeric state of a protein is harder than it looks, and discrepancies between methods are disturbingly common.
Key Takeaways
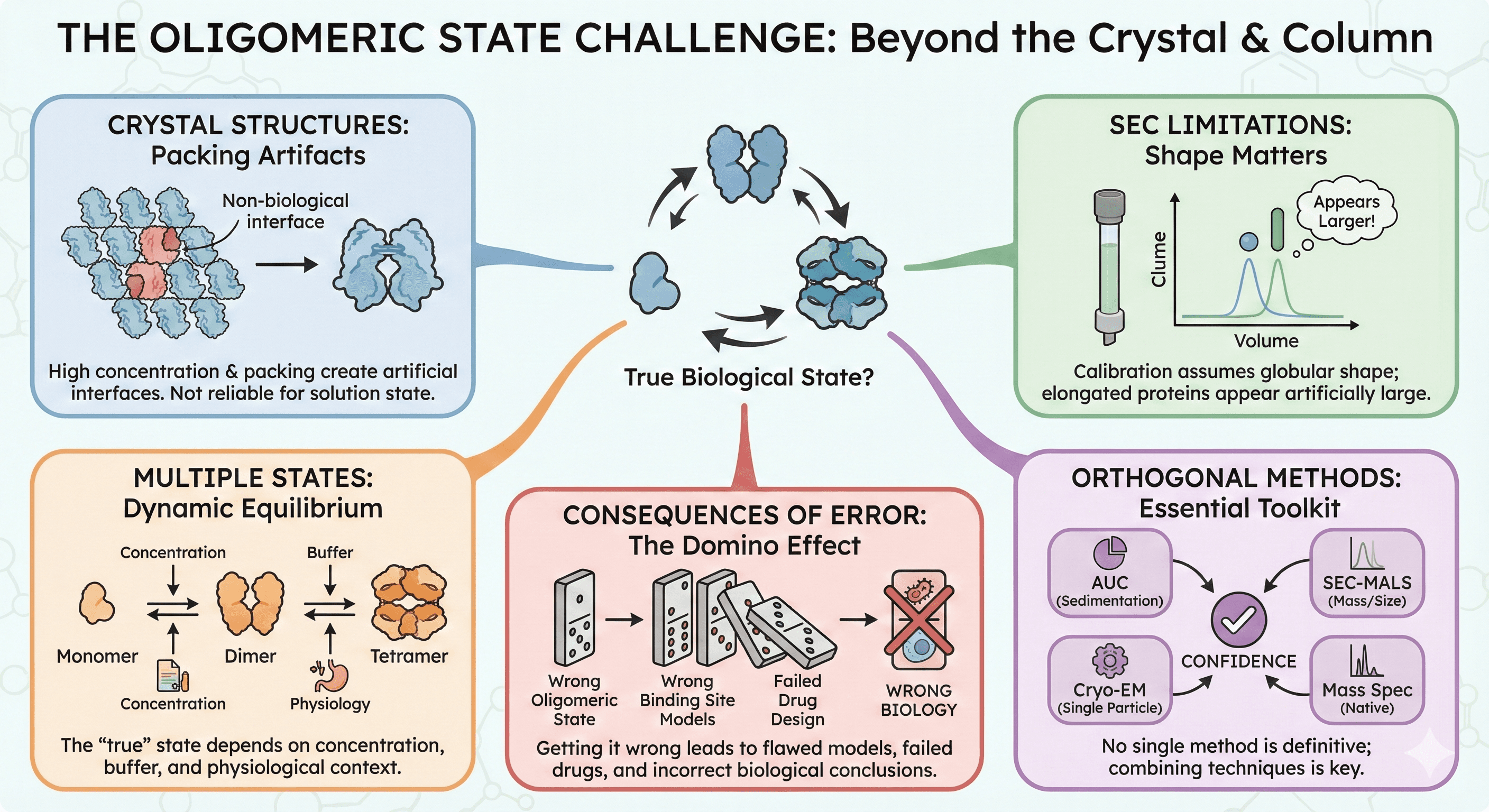
Crystal structures don't reliably report oligomeric state: High protein concentration and crystal packing create artificial interfaces
SEC is not a gold standard: Calibration curves assume globular shape; elongated proteins appear larger than they are
Most proteins exist in multiple oligomeric states: The "true" state depends on concentration, buffer, and physiological context
No single method is definitive: Orthogonal techniques are essential
Getting it wrong has consequences: Wrong oligomeric state means wrong binding site models, wrong drug design, wrong biology
The Oligomeric State Problem
Why Oligomeric State Matters
The oligomeric state determines:
Active site architecture: Many enzymes have active sites at subunit interfaces
Allosteric regulation: Cooperativity requires multi-subunit assemblies
Binding stoichiometry: A dimer has different binding properties than a monomer
Drug design: Targeting interfaces requires knowing what interfaces exist
Physiological function: Oligomers and monomers often have different functions
Get the oligomeric state wrong, and everything downstream is suspect.
The Measurement Challenge
Every method for determining oligomeric state has limitations:
Method | Measures | Assumption | Problem |
|---|---|---|---|
SEC | Elution volume | Globular shape | Elongated proteins look bigger |
Crystal structure | Lattice contacts | Biological vs. crystal contact | Hard to distinguish |
DLS | Hydrodynamic radius | Single species | Averages over mixtures |
AUC | Sedimentation coefficient | Homogeneous sample | Requires expertise |
Native MS | Mass | Survives ionization | Gas-phase artifacts |
Crosslinking | Proximity | Specific crosslinking | Non-specific crosslinks |
The Crystal Structure Problem
Why Crystals Lie About Oligomeric State
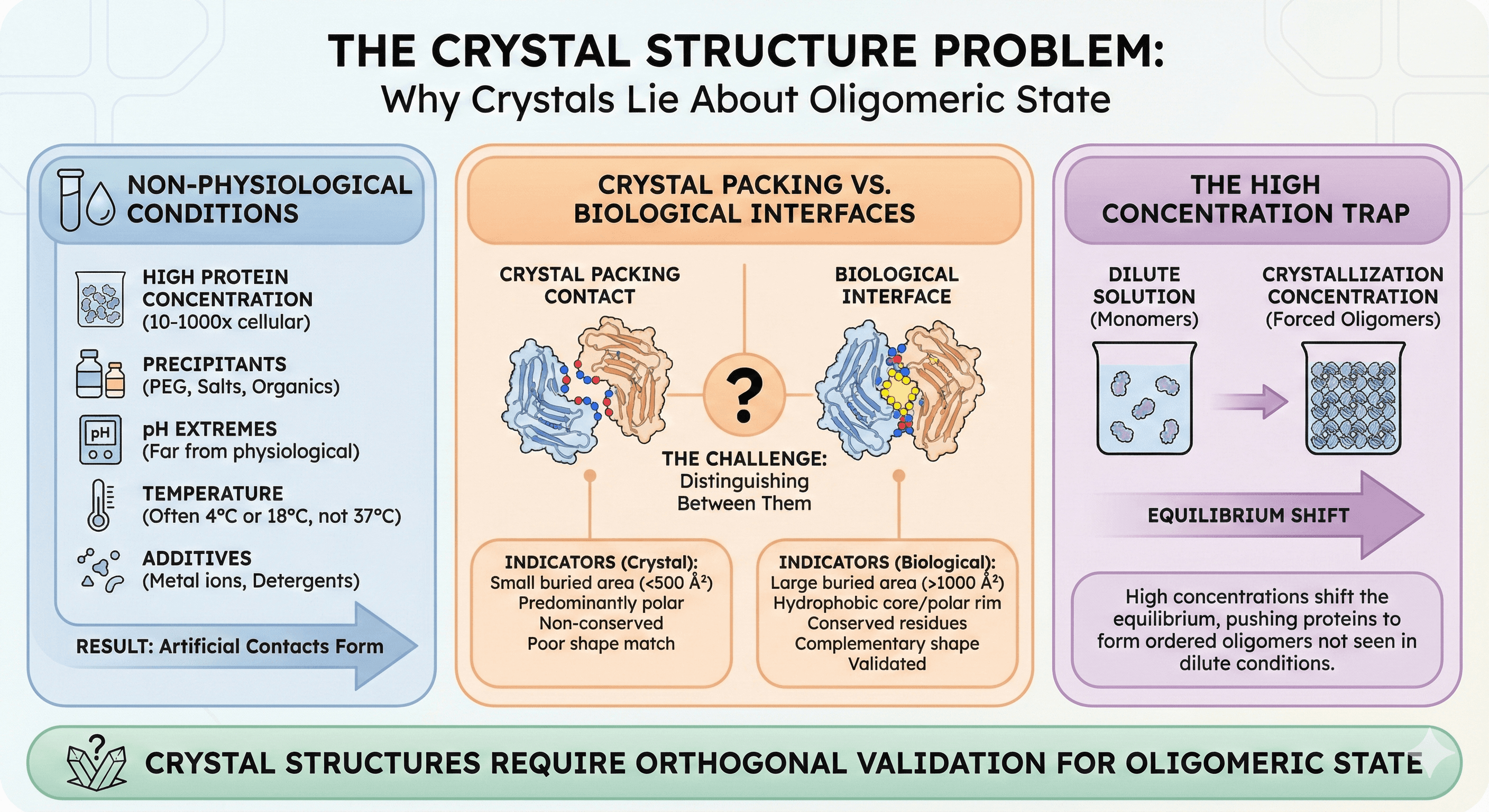
Crystallization conditions are non-physiological:
High protein concentration: 10-50 mg/mL, often 100-1000× higher than cellular
Precipitants: PEG, salts, organic solvents alter interactions
pH extremes: Many crystals grow at pH values far from physiological
Temperature: Often 4°C or 18°C, not 37°C
Additives: Metal ions, detergents can bridge subunits
The result: Proteins form contacts in crystals that don't exist in solution.
Crystal Packing vs. Biological Interfaces
Every protein crystal has multiple protein-protein contacts—that's what makes it a crystal. Studies have shown that "artificially large oligomers may be incorrectly deduced from examination of the protein–protein contacts in the crystalline environment: many of these interactions are nonspecific and simply reflect facile ways of arranging the macromolecule in a regularly ordered lattice."
The challenge: Distinguishing biological interfaces from crystal-packing contacts.
Indicators of biological interfaces:
Large buried surface area (>1000 Ų)
Hydrophobic core with polar rim
Conserved residues at interface
Complementary shape
Interface validated by other methods
Indicators of crystal contacts:
Small buried surface area (<500 Ų)
Predominantly polar
Non-conserved residues
Poor shape complementarity
Not seen in other crystal forms
The High Concentration Trap
Research on oligomeric states has demonstrated that "crystallography has limitations in determining quaternary structure, as crystallization conditions optimize for perfect order, which is achieved at high protein concentrations and solution additives (salt and crowders). These may push the proteins to form an ordered, homogeneous lattice."
Proteins that are monomers in dilute solution may form dimers or higher oligomers at crystallization concentrations simply because the equilibrium shifts.
The SEC Problem
Why SEC Lies About Oligomeric State
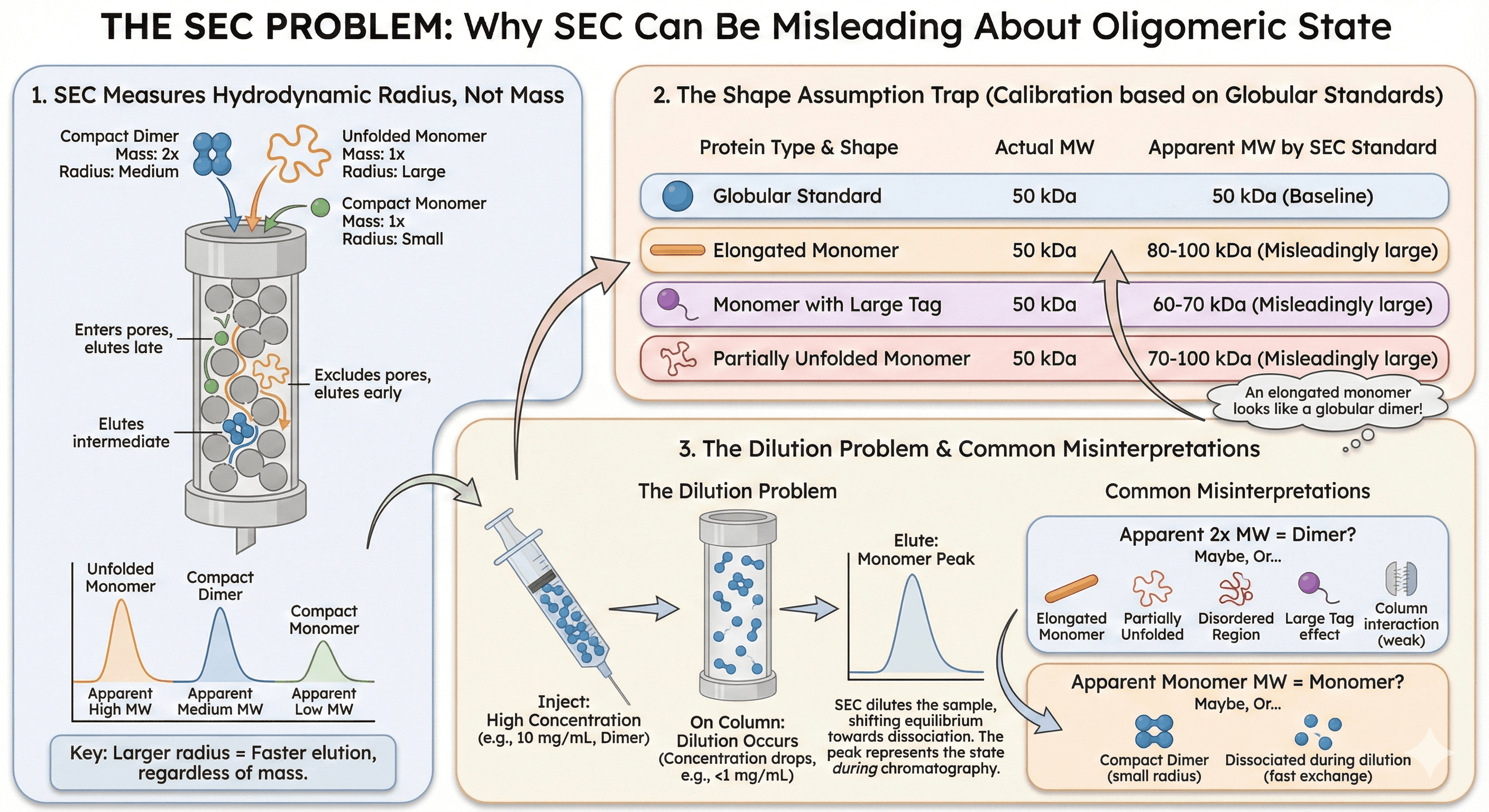
Size exclusion chromatography is the most common method for assessing oligomeric state. It's fast, easy, and gives a clear answer.
The problem: SEC doesn't measure mass—it measures hydrodynamic radius.
The Shape Assumption
SEC calibration uses globular protein standards. This creates systematic errors:
Protein Shape | Actual MW | Apparent MW by SEC |
|---|---|---|
Globular | 50 kDa | 50 kDa |
Elongated | 50 kDa | 80-100 kDa |
With large tag | 50 kDa | 60-70 kDa |
Partially unfolded | 50 kDa | 70-100 kDa |
An elongated monomer looks like a globular dimer.
Common Misinterpretations
"My protein runs at 2× the expected molecular weight—it must be a dimer"
Maybe. Or:
It's an elongated monomer
It's partially unfolded
It has a disordered region that increases hydrodynamic radius
The tag adds more volume than expected
It interacts weakly with the column matrix
"My protein runs at exactly the expected monomer MW—it's definitely a monomer"
Maybe. Or:
It's a compact dimer with half the expected hydrodynamic radius
Dimer dissociates during chromatography due to dilution
The dimer is in fast exchange and you're seeing average behavior
The Dilution Problem
SEC dilutes the sample as it passes through the column. For proteins with concentration-dependent oligomerization:
Inject: 10 mg/mL (dimer)
On column: <1 mg/mL (monomer)
Elute: monomer peak
The SEC tells you about the oligomeric state during chromatography, not before it.
The Reality: Multiple States Coexist
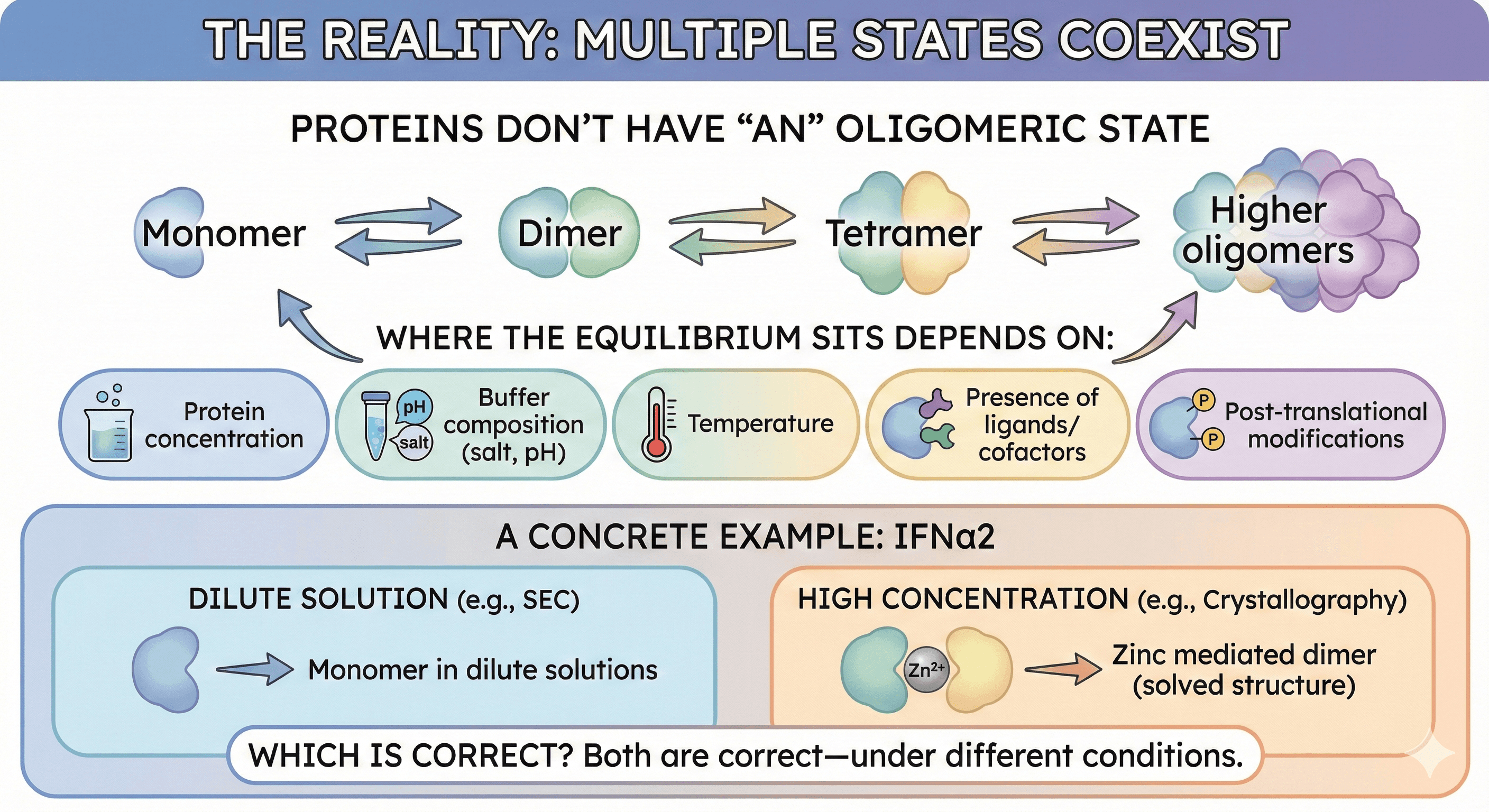
Proteins Don't Have "An" Oligomeric State
A systematic study comparing 17 proteins using multiple methods found that "it would be wrong to assign a single oligomeric state to proteins. Most proteins appear in more than one state. Moreover, of the selected 17 proteins, none is solely in a monomeric state at all protein concentrations."
The equilibrium:
Where the equilibrium sits depends on:
Protein concentration
Buffer composition (salt, pH)
Temperature
Presence of ligands/cofactors
Post-translational modifications
A Concrete Example: IFNα2
Research showed that "the SEC elution volume of IFNα2 is reduced at higher protein-concentrations, suggesting a higher oligomeric state at higher protein concentration. Indeed, while IFNα2 is a monomer in dilute solutions, it was solved as a zinc mediated dimer by X-ray crystallography."
Which is correct? Both are correct—under different conditions.
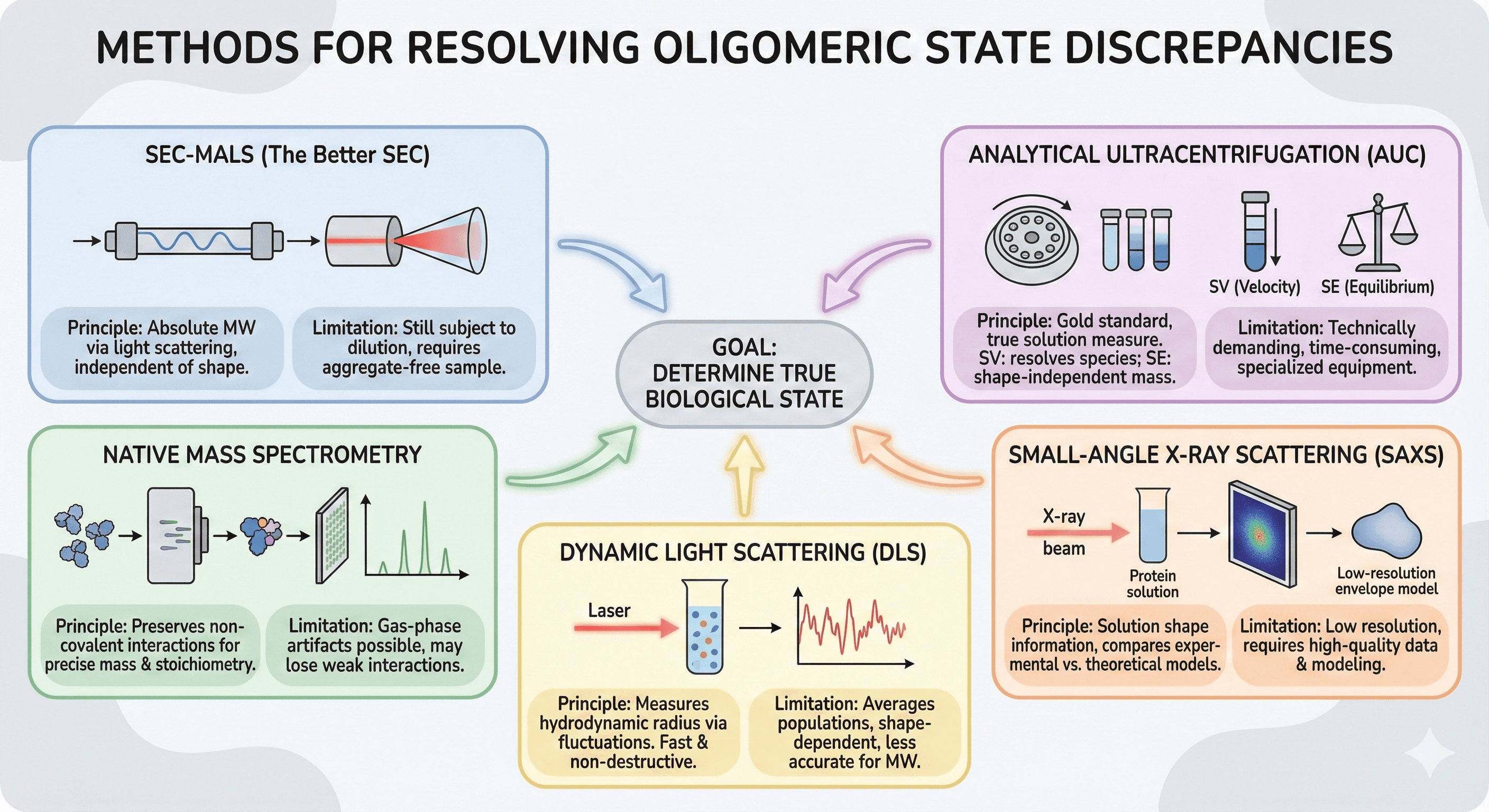
Methods for Resolving Discrepancies
SEC-MALS (The Better SEC)
Multi-angle light scattering (MALS) coupled to SEC provides absolute molecular weight, independent of shape.
Advantages:
Directly measures MW from light scattering
Doesn't require calibration standards
Works for non-globular proteins
Provides polydispersity information
Limitations:
Requires clean, aggregate-free samples
Still subject to dilution effects
Higher oligomers may dissociate on column
Rule of thumb: Designed proteins with measured oligomeric mass within ≤13% of expected are considered validated.
Analytical Ultracentrifugation (AUC)
AUC remains the gold standard for rigorous oligomeric state determination.
Sedimentation velocity (SV): Resolves species by sedimentation coefficient, allowing clear separation and quantification of monomers, oligomers, and aggregates in solution.
Sedimentation equilibrium (SE): Directly measures macromolecular mass independent of shape, making it "the method of choice for molar mass determinations and the study of self-association."
Advantages:
True solution measurement (no matrix interaction)
Can characterize equilibria between oligomeric states
Shape-independent mass determination (SE)
High resolution (SV)
Limitations:
Requires specialized equipment
Technically demanding
Time-consuming (hours to days)
Limited throughput
Native Mass Spectrometry
Native MS preserves non-covalent interactions, allowing direct determination of oligomeric state.
Advantages:
Precise mass measurement
Can resolve heterogeneous populations
Stoichiometry directly determined
Works with small sample amounts
Limitations:
Requires specialized instrumentation
Gas-phase artifacts possible
May not preserve weak interactions
Membrane proteins challenging
Small-Angle X-ray Scattering (SAXS)
SAXS provides low-resolution structural information in solution.
For oligomeric state: The quaternary structure can be deduced by comparing experimental SAXS curves to theoretical curves calculated from proposed models. This approach is especially robust when the crystal structure is known.
Advantages:
Solution measurement at relevant concentrations
Provides shape information
Can model flexible regions
Works with most proteins
Limitations:
Low resolution
Requires high-quality data
Heterogeneous samples problematic
Data interpretation requires modeling
Dynamic Light Scattering (DLS)
DLS measures hydrodynamic radius in solution.
Advantages:
Fast (minutes)
Non-destructive
Small sample volume
Good for detecting aggregation
Limitations:
Averages over populations
Can't resolve similar-sized species
Shape-dependent
Less accurate for MW determination
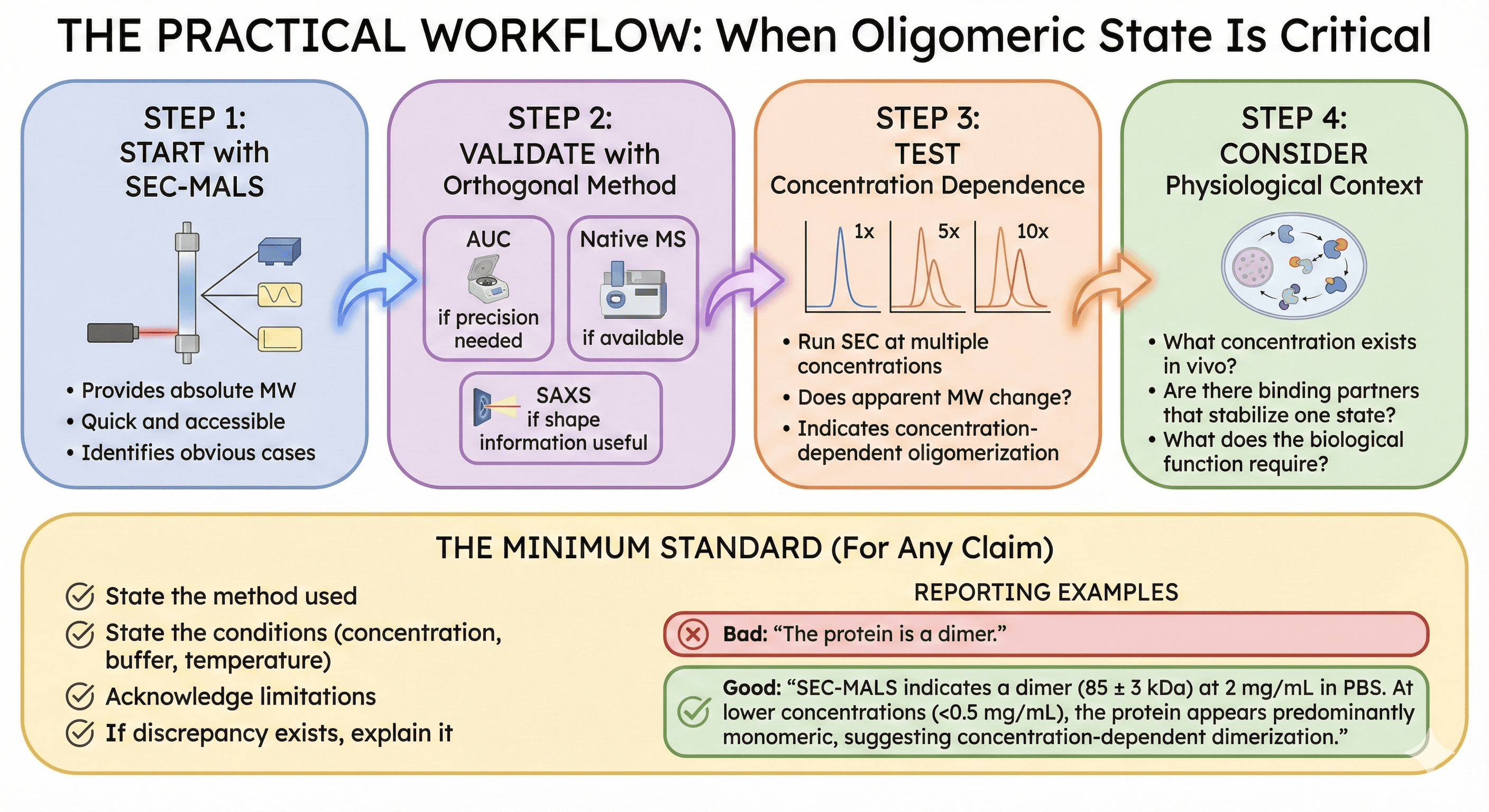
The Practical Workflow
When Oligomeric State Is Critical
Step 1: Start with SEC-MALS
Provides absolute MW
Quick and accessible
Identifies obvious cases
Step 2: Validate with orthogonal method
AUC if precision needed
Native MS if available
SAXS if shape information useful
Step 3: Test concentration dependence
Run SEC at multiple concentrations
Does apparent MW change?
This indicates concentration-dependent oligomerization
Step 4: Consider physiological context
What concentration exists in vivo?
Are there binding partners that stabilize one state?
What does the biological function require?
The Minimum Standard
For any claim about oligomeric state:
State the method used
State the conditions (concentration, buffer, temperature)
Acknowledge limitations
If discrepancy exists, explain it
Bad: "The protein is a dimer" Good: "SEC-MALS indicates a dimer (85 ± 3 kDa) at 2 mg/mL in PBS. At lower concentrations (<0.5 mg/mL), the protein appears predominantly monomeric, suggesting concentration-dependent dimerization."
Case Studies
Case 1: The Crystallographic Artifact
Protein: Metabolic enzyme from bacteria Crystal structure: Hexameric ring SEC: Monomer (45 kDa) SEC-MALS: Monomer (47 kDa) AUC: Monomer at all tested concentrations
Resolution: The hexamer was a crystallographic artifact. The six-fold symmetry of the space group promoted hexameric packing. Solution methods consistently showed monomer.
Consequence: Early drug design targeted the hexameric interface—which doesn't exist.
Case 2: The Concentration-Dependent Oligomer
Protein: Signaling protein Crystal structure: Dimer (solved at 20 mg/mL) SEC: Monomer (at 1 mg/mL) SEC-MALS at 10 mg/mL: Dimer
Resolution: Genuine concentration-dependent dimerization. Kd ≈ 5 mg/mL. Monomer and dimer both physiologically relevant at different cellular concentrations.
Consequence: Both forms studied separately; dimer has regulatory function.
Case 3: The Misleading SEC Peak
Protein: Transcription factor with long disordered N-terminus SEC: 120 kDa apparent (expected 60 kDa) Conclusion: "Must be a dimer" SEC-MALS: 62 kDa AUC: Monomer with f/f₀ = 1.8 (highly elongated)
Resolution: The disordered region increases hydrodynamic radius. The protein is a monomer that appears dimeric on calibrated SEC.
Consequence: Binding stoichiometry recalculated; mechanism revised.
Case 4: Multiple Conformations Hidden in Crystal
Protein: Metabolic enzyme (SDR family) Crystal structures: All show D2 symmetric tetramer Cryo-EM: ~50% symmetric tetramer, ~50% alternative conformations
Resolution: "The failure to observe conformations of this kind in the extensive crystallographic studies of SDR superfamily enzymes suggests that the requirement for stable packing in a regular lattice may have suppressed observation of these conformational states."
Consequence: Understanding of enzyme mechanism revised to include conformational heterogeneity.
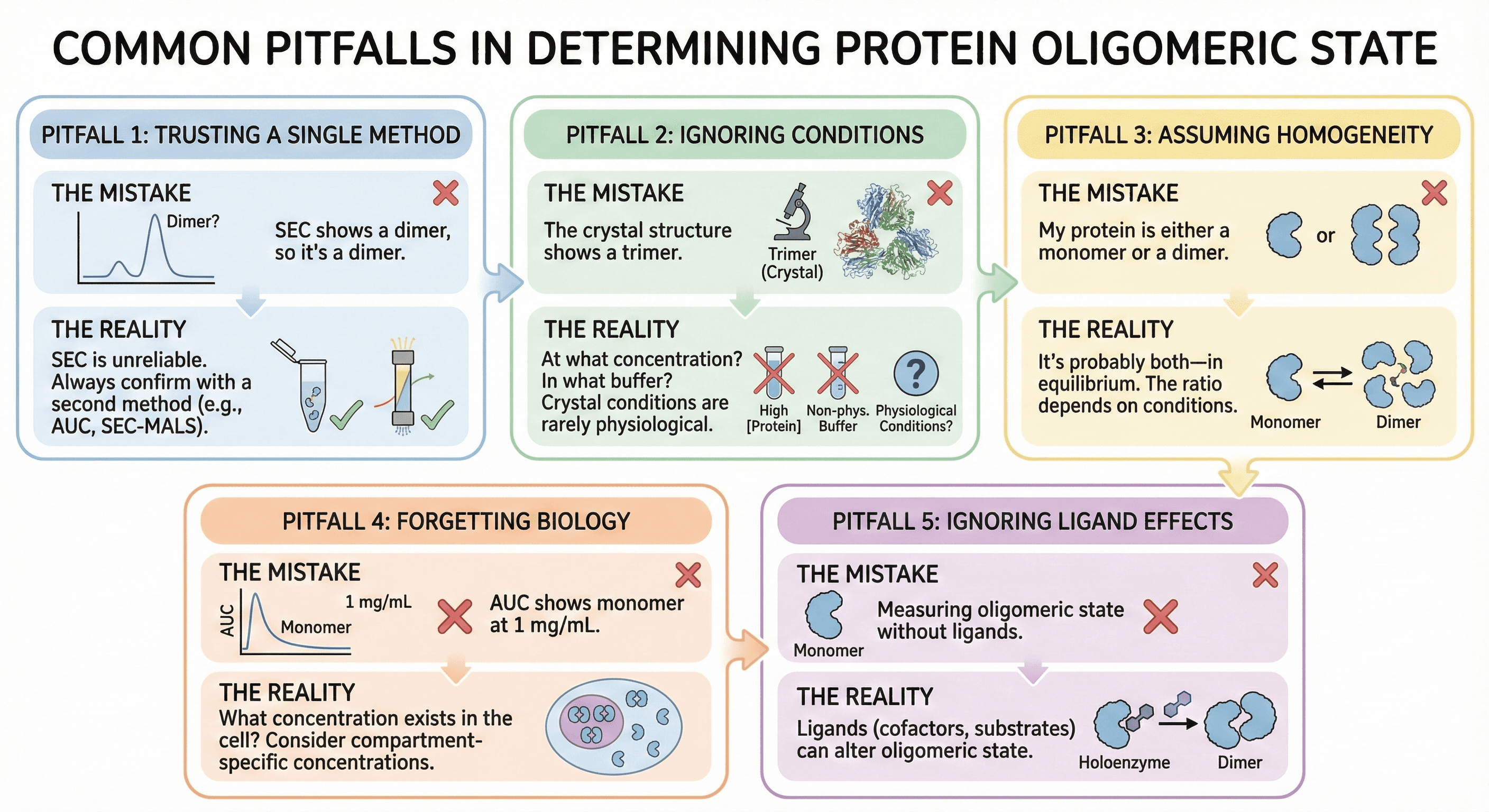
Common Pitfalls
Pitfall 1: Trusting a Single Method
The mistake: "SEC shows a dimer, so it's a dimer"
The reality: SEC is particularly unreliable for oligomeric state. Always confirm with a second method.
Pitfall 2: Ignoring Conditions
The mistake: "The crystal structure shows a trimer"
The reality: At what concentration? In what buffer? With what additives? Crystal conditions are rarely physiological.
Pitfall 3: Assuming Homogeneity
The mistake: "My protein is either a monomer or a dimer"
The reality: It's probably both—in equilibrium. The ratio depends on conditions.
Pitfall 4: Forgetting Biology
The mistake: "AUC shows monomer at 1 mg/mL"
The question: What concentration exists in the cell? If cellular concentration is 0.1 mg/mL, you've measured the relevant state. If it's 10 mg/mL (in some compartments), you may have missed the physiological oligomer.
Pitfall 5: Ignoring Ligand Effects
The mistake: Measuring oligomeric state without physiological ligands
The reality: Cofactors, substrates, and allosteric effectors can dramatically alter oligomeric state. Apoenzyme may be monomer; holoenzyme may be dimer.
The Interpretation Framework
Questions to Ask
What does the biology require?
Does function require multimerization?
Are there interface residues implicated in disease?
Do known regulatory mechanisms involve oligomerization?
Are the methods consistent?
If SEC and crystal agree: Probably correct
If methods disagree: More investigation needed
If concentration changes the answer: Concentration-dependent equilibrium
What about conservation?
Is the interface conserved across species?
Are interface residues under selection pressure?
Conserved interfaces are more likely biological
What about mutations?
Do mutations at the interface affect function?
Can you disrupt or stabilize the interface?
Mutagenesis provides functional validation
The Bottom Line
There's no simple answer to "what's the oligomeric state of my protein?" because:
Factor | Effect |
|---|---|
Concentration | Higher = more oligomer |
Buffer conditions | Can stabilize or destabilize interfaces |
Temperature | Affects equilibrium |
Ligands | Often shift equilibrium |
Method | Each has systematic biases |
The practical approach:
Use multiple methods
Test concentration dependence
Consider physiological relevance
Validate with mutagenesis if critical
The honest answer: "My protein exists as a monomer-dimer equilibrium with Kd of approximately X mg/mL, as determined by SEC-MALS and confirmed by AUC. At physiological concentrations, the predominant species is likely Y."
Structural Analysis for Oligomeric State
For researchers trying to understand their protein's quaternary structure, platforms like Orbion can provide initial guidance:
Interface analysis from predicted structures: Identify potential oligomerization surfaces
Conservation mapping: Highly conserved surface patches may indicate biological interfaces
Comparison to homologs: Do related proteins oligomerize?
Binding site prediction: Sites at putative interfaces suggest functional relevance
While computational analysis can't replace experimental validation, it can prioritize which interfaces to test and which methods to apply—helping you design the experiments that will resolve the discrepancy.
References
Krissinel E & Henrick K. (2007). Inference of macromolecular assemblies from crystalline state. Journal of Molecular Biology, 372(3):774-797. PMC2271157
Frenkel D, et al. (2022). Protein quaternary structures in solution are a mixture of multiple forms. Chemical Science, 13:11687-11701. PMC9555727
Lebowitz J, et al. (2002). Modern analytical ultracentrifugation in protein science: A tutorial review. Protein Science, 11(9):2067-2079. PMC2373601
Schuck P. (2016). Analytical ultracentrifugation in structural biology. Progress in Biophysics and Molecular Biology, 129:54-76. PMC5899701
Putnam CD, et al. (2007). X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Quarterly Reviews of Biophysics, 40(3):191-285. PMC5866936
Keifer DZ & Pierson EE. (2023). Native mass spectrometry: Recent progress and remaining challenges. Annual Review of Analytical Chemistry, 16:475-495. PMC10700022
Akey DL, et al. (2022). Oligomeric interactions maintain active-site structure in a noncooperative enzyme family. Protein Science, 31(9):e4398. PMC9433937
Slabinski L, et al. (2007). The challenge of protein structure determination—lessons from structural genomics. Protein Science, 16(11):2472-2482.

