Blog
Why Your Protein Aggregates (It's Missing Zinc)
Dec 17, 2025
You purified your enzyme. The gel looks perfect—a single band at the expected molecular weight. SEC shows a beautiful monomer peak. Then you run the activity assay, and... nothing. Zero activity. A week later, you notice your protein has crashed out of solution—white precipitate at the bottom of the tube.
What happened? Your protein lost its cofactor during purification. Without it, the active site collapsed, exposed hydrophobic residues aggregated, and now you have expensive, inactive precipitate instead of functional enzyme.
This is one of the most underappreciated failure modes in protein science. Here's why cofactors matter, how they're lost, and the devastating consequences when they disappear.
Key Takeaways
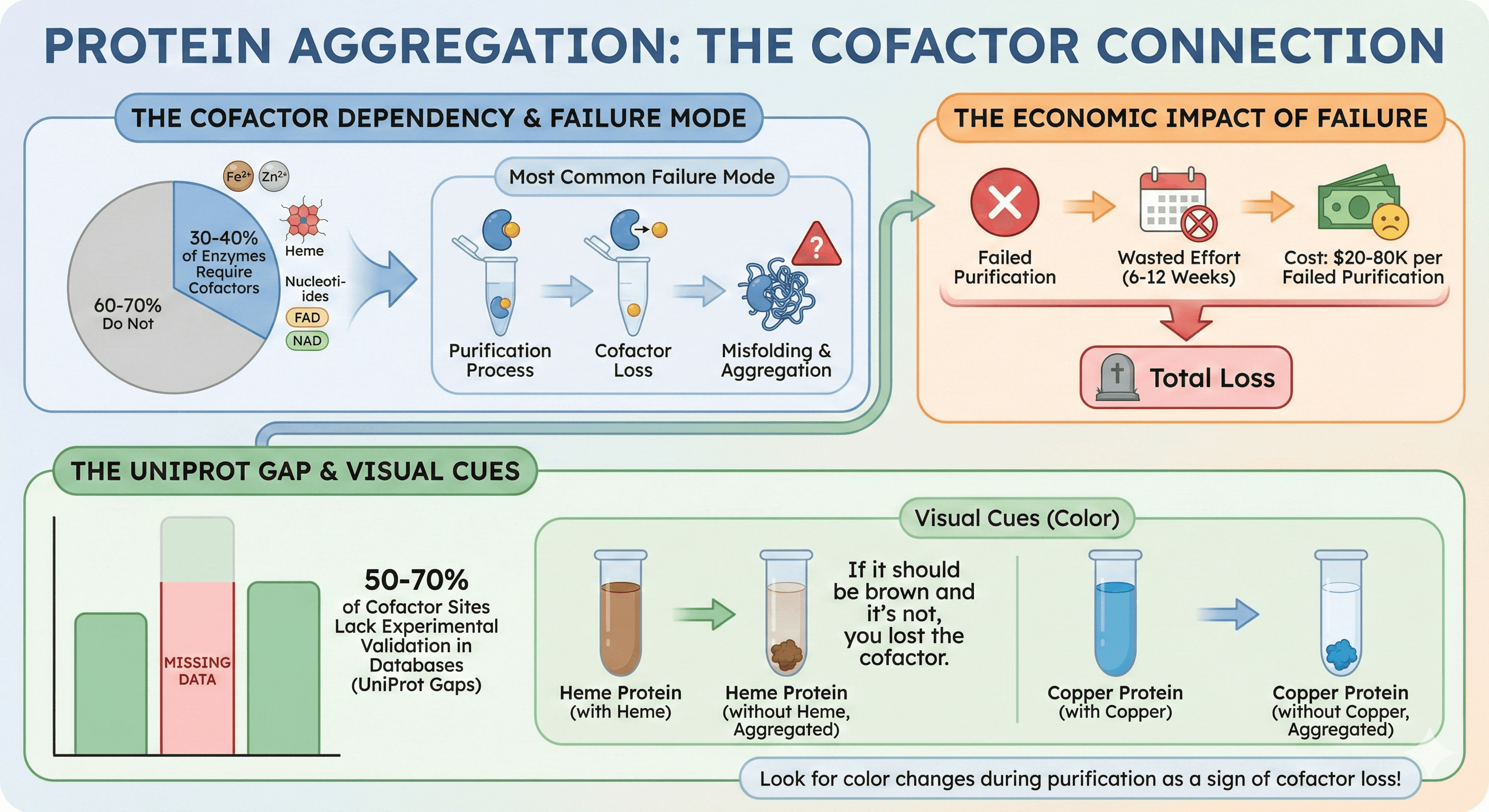
30-40% of all enzymes require cofactors (metal ions, nucleotides, heme, FAD/NAD) for function or stability
Most common failure mode: Cofactor loss during purification → misfolding → aggregation
Economic impact: $20-80K per failed purification (6-12 weeks of wasted effort)
The problem: 50-70% of cofactor sites lack experimental validation in databases (UniProt gaps)
Visual cues: If your protein should be colored (heme = brown, Fe-S = brown, copper = blue) and it's not, you lost the cofactor
What Are Cofactors?
Cofactors are non-protein molecules that bind to proteins and are essential for function, stability, or both.
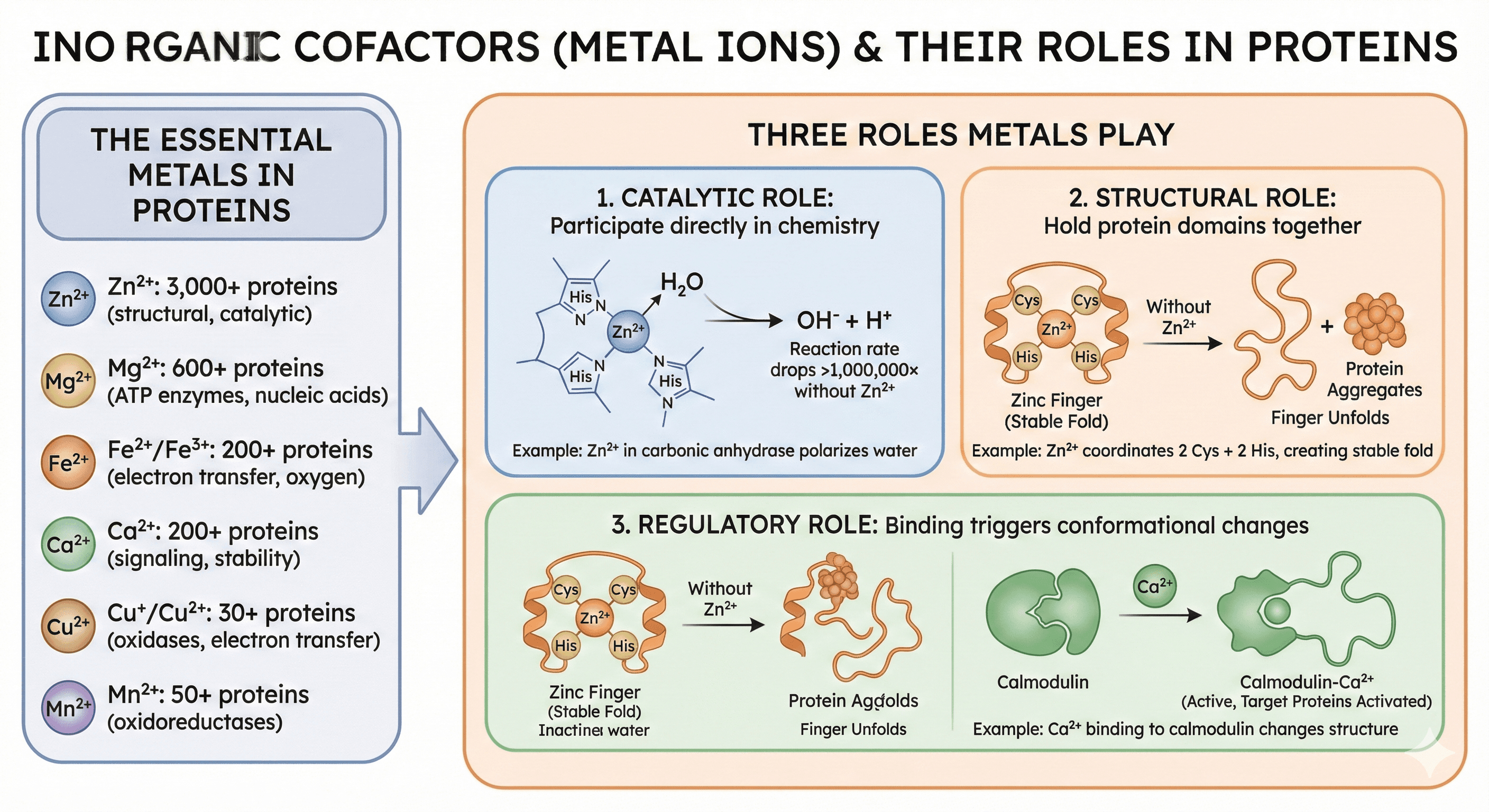
Inorganic Cofactors (Metal Ions)
The essential metals in proteins:
Zinc (Zn²⁺): 3,000+ human proteins (most abundant metal in structural proteins)
Magnesium (Mg²⁺): 600+ proteins (ATP-dependent enzymes, nucleic acid binding)
Iron (Fe²⁺/Fe³⁺): 200+ proteins (electron transfer, oxygen binding)
Calcium (Ca²⁺): 200+ proteins (signaling, structural stability)
Copper (Cu⁺/Cu²⁺): 30+ proteins (oxidases, electron transfer)
Manganese (Mn²⁺): 50+ proteins (oxidoreductases)
Three roles metals play:
1. Catalytic: Participate directly in chemistry
Example: Zn²⁺ in carbonic anhydrase polarizes water (makes H₂O easier to deprotonate)
Without Zn²⁺: Reaction rate drops >1,000,000×
2. Structural: Hold protein domains together
Example: Zn²⁺ in zinc fingers coordinates 2 Cys + 2 His, creating stable fold
Without Zn²⁺: Finger unfolds within seconds, protein aggregates
3. Regulatory: Binding triggers conformational changes
Example: Ca²⁺ binding to calmodulin changes structure, activates target proteins
Organic Cofactors (Coenzymes)
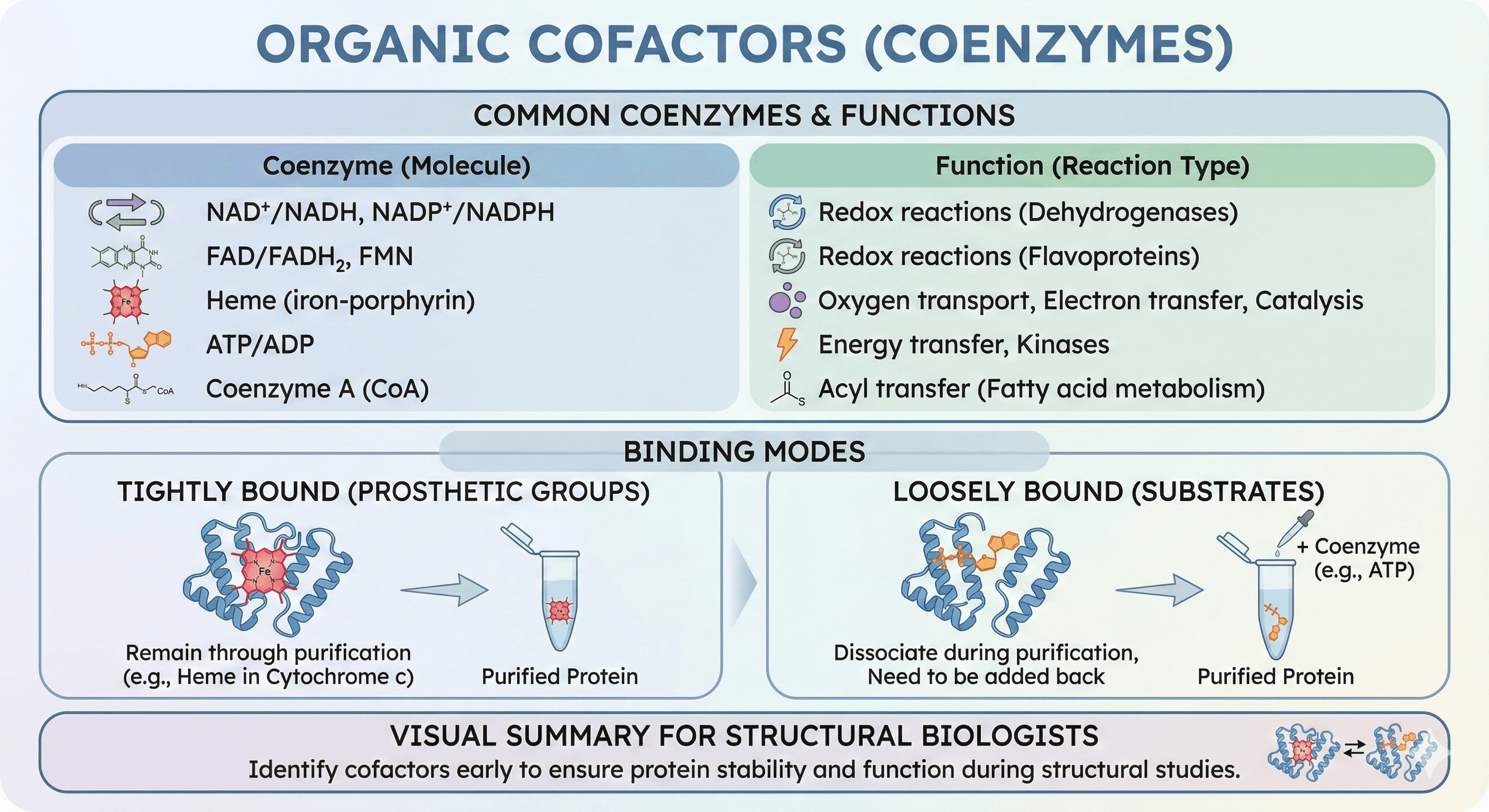
Common coenzymes:
NAD⁺/NADH, NADP⁺/NADPH: Redox reactions (dehydrogenases)
FAD/FADH₂, FMN: Redox reactions (flavoproteins)
Heme (iron-porphyrin): Oxygen transport, electron transfer, catalysis
ATP/ADP: Energy transfer, kinases
Coenzyme A (CoA): Acyl transfer (fatty acid metabolism)
Binding modes:
Tightly bound (prosthetic groups): Remain through purification (e.g., heme in cytochrome c)
Loosely bound (substrates): Dissociate during purification, need to be added back
The Three Critical Roles of Cofactors
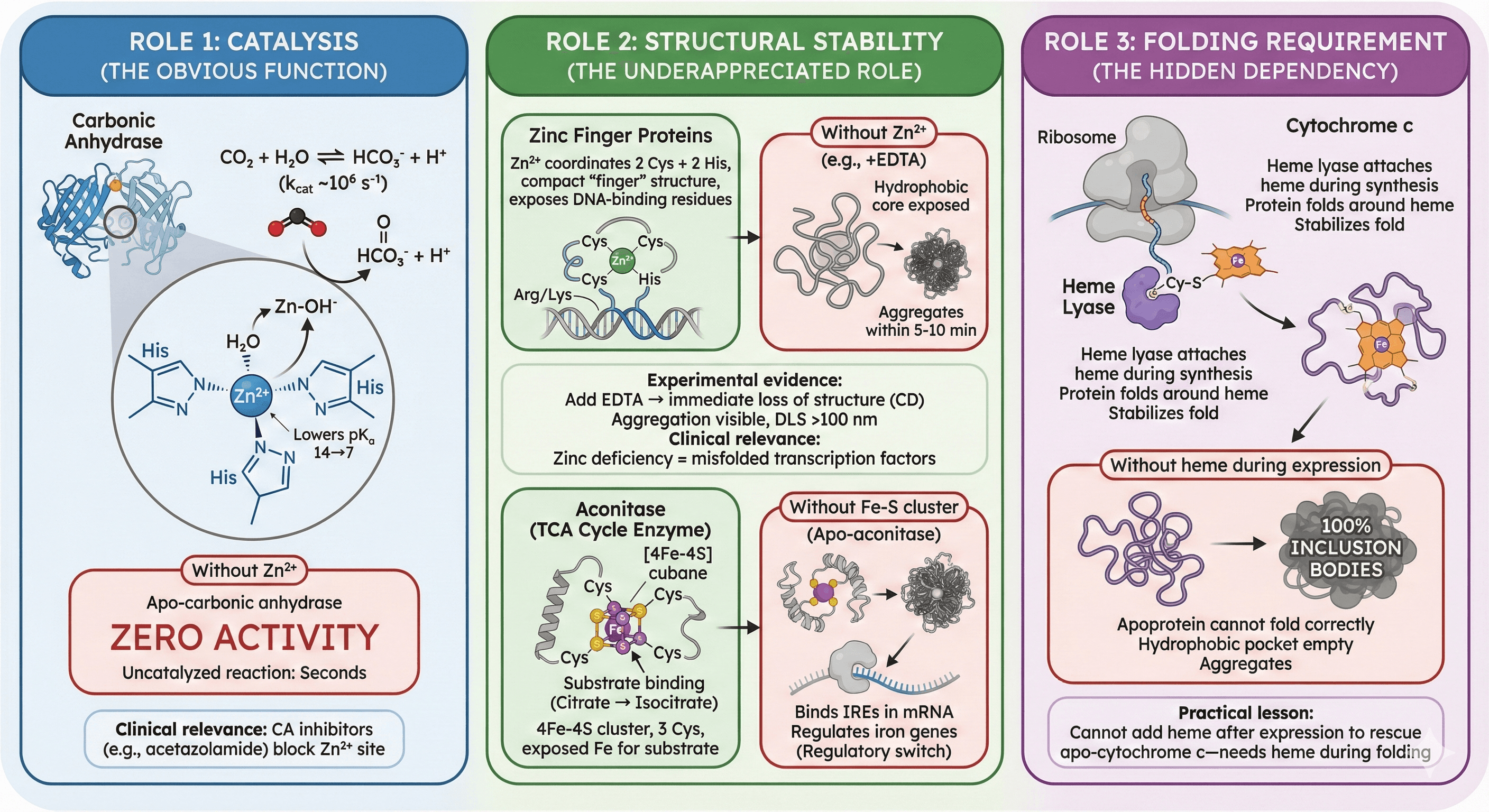
Role 1: Catalysis (The Obvious Function)
Example: Carbonic Anhydrase
Function: Catalyzes CO₂ + H₂O ⇌ HCO₃⁻ + H⁺ (one of the fastest enzymes, kcat ~10⁶ s⁻¹)
Cofactor: Zn²⁺ at the active site
Mechanism:
Zn²⁺ coordinates 3 His residues + 1 H₂O
Zn²⁺ polarizes water, lowering pKa from 14 → 7
Zn-OH⁻ nucleophile attacks CO₂, forming bicarbonate
Without Zn²⁺:
Apo-carbonic anhydrase: Zero activity (kcat drops >1,000,000×)
Uncatalyzed reaction: Takes seconds (vs microseconds with enzyme)
Clinical relevance: CA inhibitors (acetazolamide) treat glaucoma by blocking Zn²⁺ site
Role 2: Structural Stability (The Underappreciated Role)
This is where people get surprised. Even if the cofactor isn't directly involved in catalysis, its loss causes misfolding or aggregation.
Example: Zinc Finger Proteins
Structure: Classic Cys₂His₂ zinc finger
Zn²⁺ coordinates 2 Cys + 2 His residues
Coordination pulls polypeptide into compact "finger" structure
Exposes DNA-binding residues (Arg, Lys) on one face
Without Zn²⁺:
Finger unfolds within seconds
Hydrophobic core becomes exposed
Protein aggregates within 5-10 minutes
Experimental evidence:
Add EDTA (chelates Zn²⁺) → immediate loss of structure (CD spectroscopy)
Aggregation visible by eye (cloudiness) within 5-10 minutes
DLS shows particles >100 nm (aggregates)
Clinical relevance: Zinc deficiency causes immunodeficiency (zinc finger transcription factors misfold)
Example: Aconitase (TCA Cycle Enzyme)
Structure: [4Fe-4S] cubane cluster
4 iron atoms + 4 sulfur atoms in cubic arrangement
Held by 3 Cys residues
One Fe is exposed for substrate binding
Function: Converts citrate → isocitrate (TCA cycle)
Without Fe-S cluster:
Apo-aconitase loses structure around active site
Substrate-binding cleft collapses
Protein aggregates during storage
Interesting twist: Apo-aconitase becomes an RNA-binding protein (IRP1)
Acts as iron sensor: When Fe is low, cluster disassembles
Apo form binds iron-responsive elements (IREs) in mRNA
Regulates iron metabolism genes
This is a regulatory switch (holo = enzyme, apo = RNA binding)
Role 3: Folding Requirement (The Hidden Dependency)
Some proteins cannot fold without their cofactor present during translation.
Example: Cytochrome c
Structure: 12 kDa protein with covalently attached heme
Heme attached via thioether bonds to 2 Cys residues
Heme sits in hydrophobic pocket, stabilizes fold
Folding pathway:
Nascent polypeptide synthesized on ribosome
Heme lyase enzyme attaches heme (while protein still in ribosome tunnel)
Protein folds around heme
Without heme during expression:
Apoprotein cannot fold correctly
Hydrophobic pocket (meant for heme) is empty
Protein aggregates → 100% inclusion bodies
Experimental proof:
Express cytochrome c in E. coli without heme biosynthesis genes → insoluble
Co-express hem operon (heme synthesis genes) → soluble, functional protein
Practical lesson: You cannot add heme after expression to rescue apo-cytochrome c—it needs heme during folding
How Proteins Lose Their Cofactors
The Purification Gauntlet
Starting point: Cell lysate, protein contains bound cofactor (Zn²⁺, Mg²⁺, heme, etc.)
Purification steps where cofactors are lost:
1. Cell Lysis
High salt (0.5-1 M NaCl) competes with metal-protein interactions
Weakly bound metals (Mg²⁺, Ca²⁺) dissociate
Result: 20-50% metal loss in first step
2. Affinity Chromatography (His-tag)
Imidazole (250-500 mM for elution) is a metal chelator
Imidazole competes for Zn²⁺, Ni²⁺, Co²⁺
Result: Metals strip from protein during elution
This is the #1 cause of metal loss
3. Desalting (Buffer Exchange)
Dilution into metal-free buffer
Equilibrium shifts toward apo form (Le Chatelier's principle)
Result: Slow dissociation
4. Size-Exclusion Chromatography (SEC)
Long residence time in column (30-60 minutes)
Slow metal dissociation continues
Result: Further metal loss
5. Concentration
Apo-protein (cofactor-free) aggregates at high concentration
Precipitation, loss of sample
This is where you notice the problem
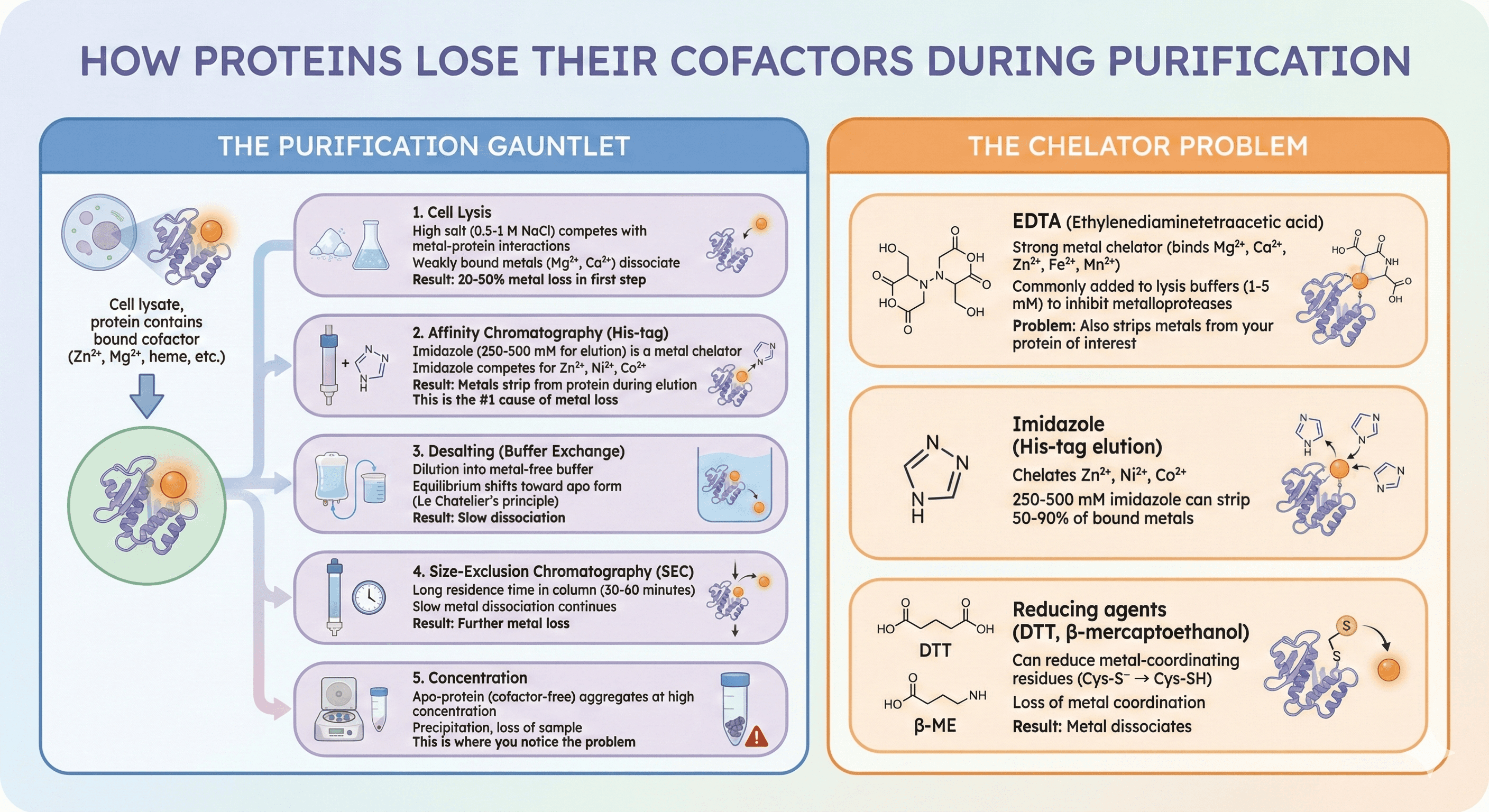
The Chelator Problem
EDTA (ethylenediaminetetraacetic acid):
Strong metal chelator (binds Mg²⁺, Ca²⁺, Zn²⁺, Fe²⁺, Mn²⁺)
Commonly added to lysis buffers (1-5 mM) to inhibit metalloproteases
Problem: Also strips metals from your protein of interest
Imidazole (His-tag elution):
Chelates Zn²⁺, Ni²⁺, Co²⁺
250-500 mM imidazole can strip 50-90% of bound metals
Reducing agents (DTT, β-mercaptoethanol):
Can reduce metal-coordinating residues (Cys-S⁻ → Cys-SH)
Loss of metal coordination
Result: Metal dissociates
Case Study 1: The Vanishing Zinc Finger
Target: Transcription factor with 4 zinc fingers (DNA-binding domain)
Expression: E. coli BL21(DE3), high yield (50 mg/L)
Purification:
Ni-NTA (His-tag), elute with 500 mM imidazole
Desalt into PBS
SEC (Superdex 75)
Result:
Initial SEC peak: Sharp monomer, looks perfect
After concentration to 5 mg/mL: 50% precipitation
After 3 days at 4°C: 90% precipitation
Investigation:
CD spectroscopy: Loss of α-helical structure over time
EMSA (DNA binding assay): No DNA binding (should retard DNA, doesn't)
ICP-MS (metal quantification): Zn content <10% of expected
Should be 4 Zn per protein
Only 0.3 Zn detected
Root cause:
Imidazole (500 mM in elution buffer) stripped Zn²⁺ from fingers
Apo-fingers unfolded, aggregated during storage
Solution:
Add 100 μM ZnCl₂ to all buffers (lysis, wash, elution, storage)
Lower imidazole concentration (500 mM → 250 mM), desalt immediately
Add 10% glycerol to storage buffer (stabilizes fold)
Result after optimization:
Zn content: 3.8 Zn per protein (>95% occupancy)
CD spectrum: Strong α-helical signal, stable for weeks
DNA binding: Restored (clean EMSA shift)
Aggregation: <5% after 2 weeks at 4°C
Time saved: 3 months (avoided project restart)
Case Study 2: The Inactive Kinase (Missing Magnesium)
Target: Ser/Thr kinase for phosphorylation studies
Expression: Insect cells (Sf9), baculovirus, 10 mg/L yield
Purification: Ni-NTA + SEC, looks clean on SDS-PAGE
Problem:
Kinase assay: Zero activity (should phosphorylate substrate, doesn't)
Autophosphorylation: None (kinases often autophosphorylate activation loop)
Crystallization: Forms crystals, but in inactive conformation (activation loop blocks active site)
Investigation:
ATP binding assay (fluorescence polarization): Binds weakly (Kd ~500 μM, should be <10 μM)
AlphaFold structure: Shows Mg²⁺ coordination site (Asp166, Asn184, conserved)
ICP-MS: Mg content <5% of expected
Root cause:
Mg²⁺ required for ATP binding (coordinates β- and γ-phosphates)
Without Mg²⁺, ATP binds 50-100× weaker
Kinase adopts inactive conformation
Solution:
Add 10 mM MgCl₂ to kinase assay buffer
Add 5 mM MgCl₂ to storage buffer
Re-test activity
Result:
Kinase activity: Fully restored (kcat/Km matches literature)
ATP binding: Kd = 5 μM (100× improvement)
Autophosphorylation: Visible (phospho-antibody detects activated kinase)
Lesson: Always check if your enzyme requires divalent cations (Mg²⁺, Mn²⁺, Ca²⁺) and add them to assay buffers
Case Study 3: The Heme-Free Peroxidase
Target: Horseradish peroxidase (HRP) for biosensor application
Expression: Pichia pastoris (secreted into media)
Purification:
Ammonium sulfate precipitation (pH 4.5)
Ion exchange (Q Sepharose)
SEC
Problem:
Enzyme is colorless (should be brown due to heme)
No peroxidase activity (should oxidize ABTS substrate → green color)
Protein looks fine on SDS-PAGE (single band, 44 kDa)
Investigation:
UV-Vis spectroscopy: No Soret band at 403 nm (characteristic of heme)
Conclusion: Heme was lost during purification (apo-HRP)
Why heme was lost:
Heme binds non-covalently in HRP (unlike cytochrome c with covalent attachment)
Acidic pH during precipitation (pH 4.5) weakened heme binding
High ionic strength (1 M (NH₄)₂SO₄) disrupted electrostatic interactions
Result: Heme dissociated, removed during SEC
Solution (heme reconstitution):
Avoid acidic pH (keep pH ≥6.5)
Add hemin (oxidized heme) during reconstitution:
Dissolve hemin in 0.1 M NaOH (heme is insoluble at neutral pH)
Dilute into protein solution (10× excess hemin)
Incubate 1 hour at 4°C
Remove excess hemin by SEC
Result:
UV-Vis: Strong Soret band at 403 nm (heme incorporated)
Activity: Fully restored (comparable to commercial HRP)
Color: Brown (heme visible)
Practical tip: If your protein should be colored and it's not, you've lost the cofactor
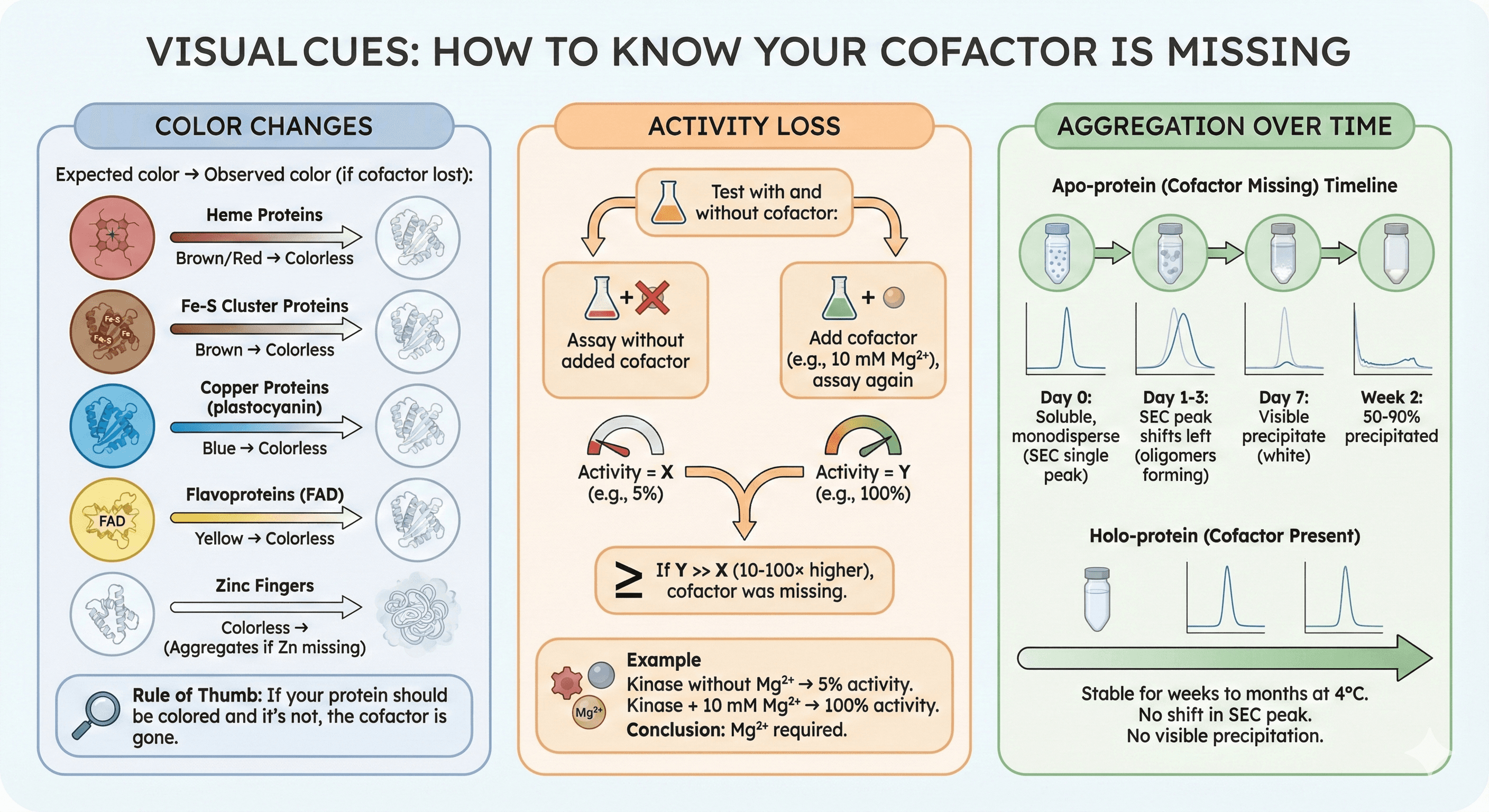
Visual Cues: How to Know Your Cofactor Is Missing
Color Changes
Expected color → Observed color (if cofactor lost):
Heme proteins: Brown/red → Colorless
Fe-S cluster proteins: Brown → Colorless
Copper proteins (plastocyanin): Blue → Colorless
Flavoproteins (FAD): Yellow → Colorless
Zinc fingers: Colorless (expected, but will aggregate if Zn missing)
Rule of thumb: If your protein should be colored and it's not, the cofactor is gone
Activity Loss
Test with and without cofactor:
Assay without added cofactor → Activity = X
Add cofactor (e.g., 10 mM Mg²⁺), assay again → Activity = Y
If Y >> X (10-100× higher), cofactor was missing
Example:
Kinase without Mg²⁺ → 5% activity
Kinase + 10 mM Mg²⁺ → 100% activity
Conclusion: Mg²⁺ required
Aggregation Over Time
Timeline of aggregation (apo-protein):
Day 0: Soluble, monodisperse (SEC shows single peak)
Day 1-3: SEC peak shifts left (oligomers forming)
Day 7: Visible precipitate (white)
Week 2: 50-90% precipitated
If cofactor is present:
Stable for weeks to months at 4°C
No shift in SEC peak
No visible precipitation
The Broader Impact: Cofactors in Biology
Why Cofactors Evolved
Metals are catalytic powerhouses:
Lewis acids (accept electrons): Zn²⁺, Mg²⁺, Fe³⁺
Redox chemistry: Fe, Cu, Mn (switch oxidation states)
Structural scaffolds: Zn²⁺ fingers, Ca²⁺ EF-hands
Coenzymes extend reaction repertoire:
NAD⁺/FAD: Redox reactions (transfer electrons)
CoA: Acyl transfer (attach fatty acids)
SAM: Methyl transfer (methylate DNA, RNA, proteins)
Evolution's solution:
Proteins alone: Limited chemical versatility (20 amino acids)
Proteins + cofactors: Expand chemistry (metals do reactions proteins can't)
Cofactor Deficiencies in Disease
Zinc deficiency:
Impaired immune function (zinc finger transcription factors misfold)
Growth retardation
Wound healing defects
Iron deficiency:
Anemia (hemoglobin needs heme)
Fatigue (cytochrome oxidase needs Fe for ATP synthesis)
Magnesium deficiency:
Muscle cramps (kinases need Mg²⁺ for ATP)
Cardiac arrhythmias
Vitamin B12 (coenzyme) deficiency:
Pernicious anemia
Neurological damage
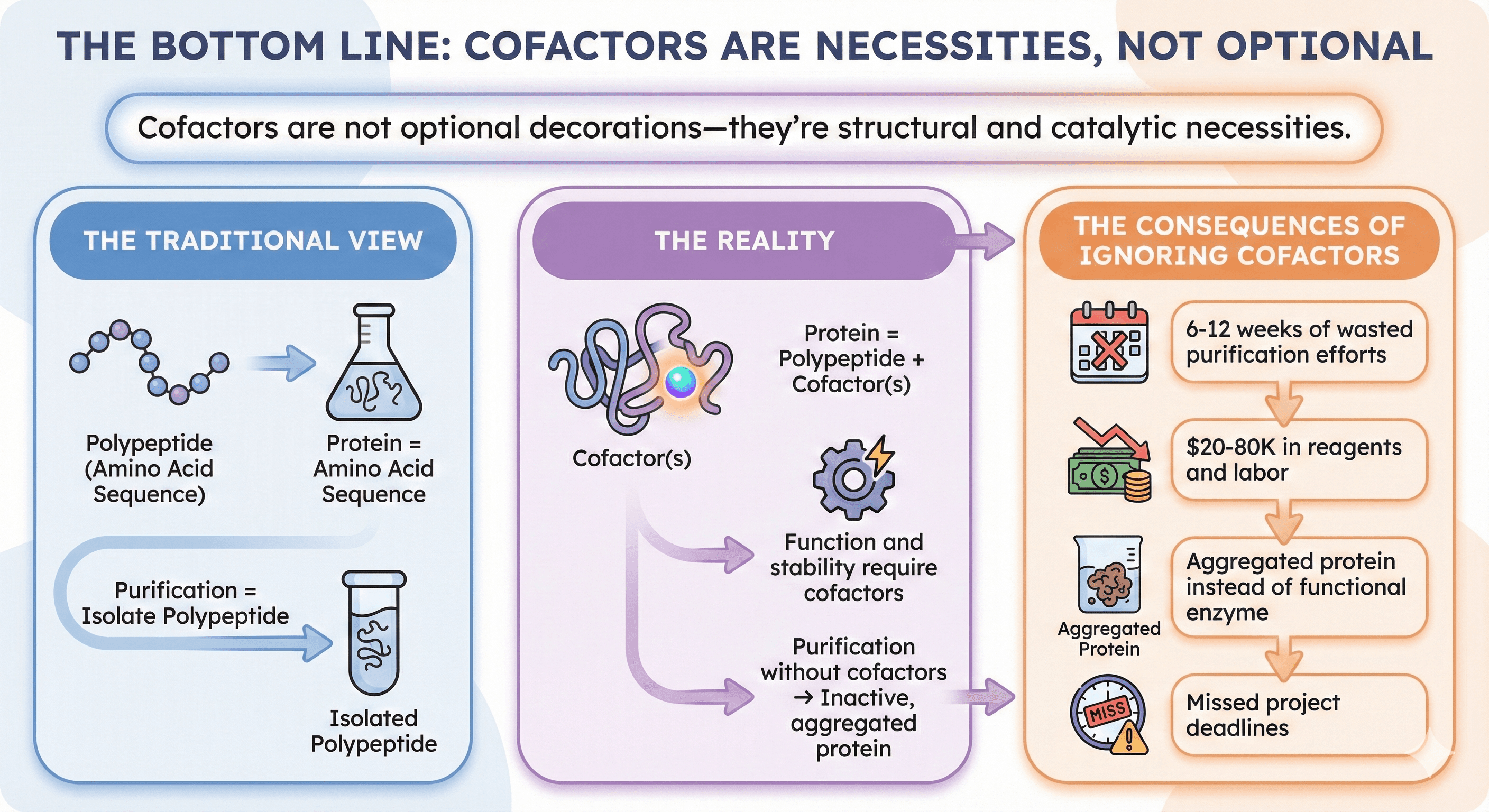
The Bottom Line
Cofactors are not optional decorations—they're structural and catalytic necessities.
The traditional view:
Protein = amino acid sequence
Purification = isolate polypeptide
The reality:
Protein = polypeptide + cofactor(s)
Function and stability require cofactors
Purification without cofactors → inactive, aggregated protein
The consequences of ignoring cofactors:
6-12 weeks of wasted purification efforts
$20-80K in reagents and labor
Aggregated protein instead of functional enzyme
Missed project deadlines
Key Takeaway
Your protein isn't broken. It's just missing its zinc.
The difference between functional enzyme and expensive precipitate is often just adding 100 μM ZnCl₂ to your buffers—if you know to look for it.

