Blog
Membrane Protein Purification: Detergent Screening Guide
Dec 19, 2025
You've spent six months optimizing expression. Finally, your Western blot shows your GPCR is in the membrane fraction—success! Now you just need to purify it. You add detergent, solubilize the membranes, run it through Ni-NTA, and... it's gone. Either it didn't bind, it aggregated during elution, or it eluted as a heterogeneous smear.
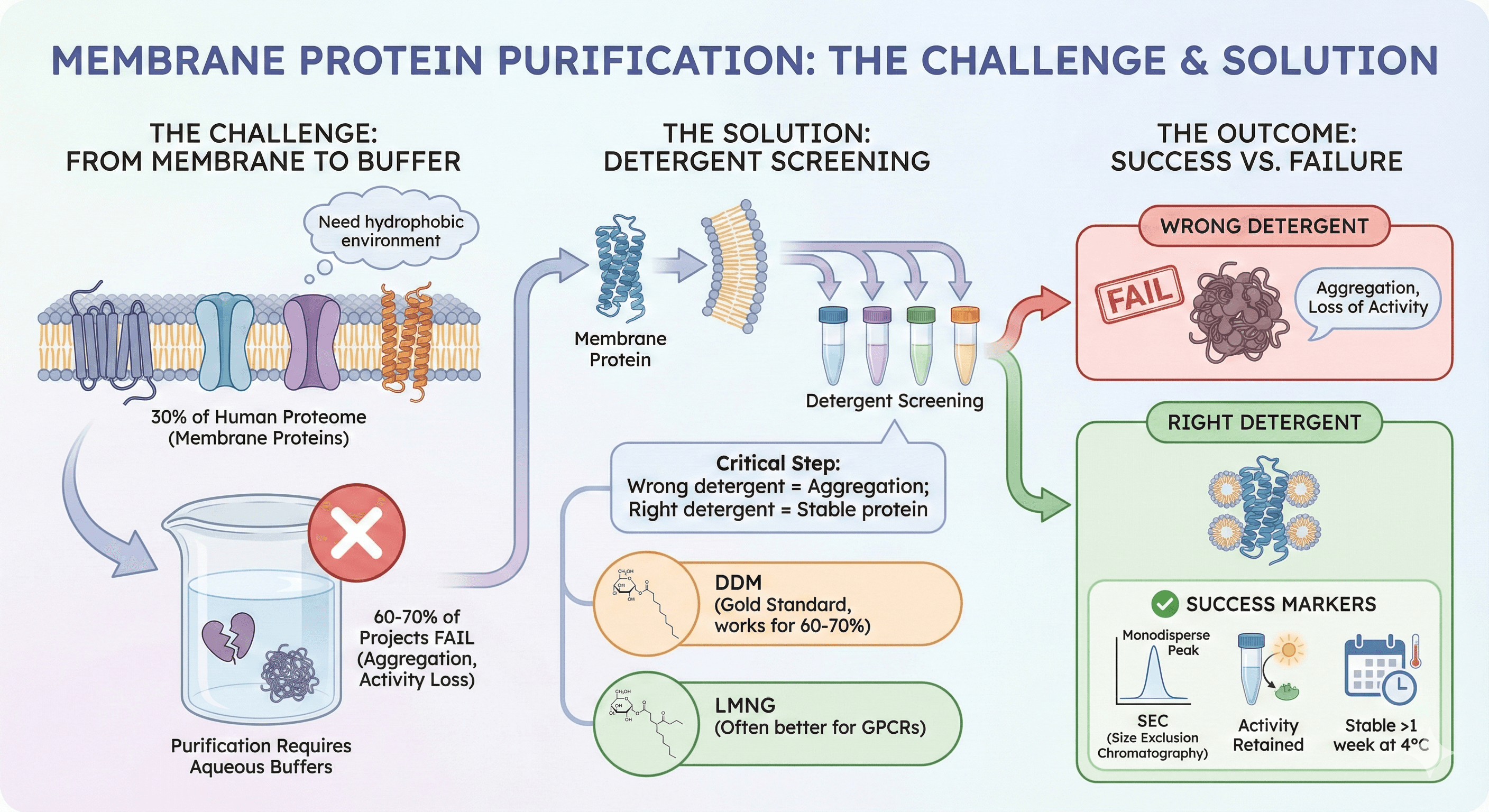
Welcome to membrane protein purification—the point where 60-70% of membrane protein projects fail. The protein is made, it's in the membrane, but you cannot get it out in a functional, stable form. Here's why membrane proteins are uniquely challenging, and how to actually solubilize and purify them.
Key Takeaways
30% of the human proteome is membrane proteins (GPCRs, ion channels, transporters)
60-70% of membrane protein projects fail during purification (aggregation, loss of activity)
The challenge: Membrane proteins need hydrophobic environment (lipid bilayer), but purification requires aqueous buffers
Detergent screening is critical: Wrong detergent = aggregation; right detergent = stable protein
DDM is gold standard (works for 60-70% of proteins), but LMNG often better for GPCRs
Success markers: SEC shows monodisperse peak, activity retained, stable >1 week at 4°C
Why Membrane Proteins Are Hard to Purify
The Fundamental Problem: Hydrophobic Surface
Soluble proteins:
Hydrophobic residues buried in core
Hydrophilic residues on surface (interact with water)
Stable in aqueous buffer
Membrane proteins:
Hydrophobic residues on surface (transmembrane helices interact with lipid tails)
Hydrophilic residues at loops (extracellular/intracellular)
Cannot exist stably in aqueous buffer (exposed hydrophobic surface drives aggregation)
The dilemma:
In membrane: Protein is stable, functional, but cannot purify (embedded in lipid bilayer)
In aqueous buffer: Can purify, but protein aggregates (exposed hydrophobic surface)
Solution: Introduce amphipathic molecules (detergents, nanodiscs, amphipols) that:
Shield hydrophobic transmembrane regions
Provide solubility in aqueous buffer
Maintain protein stability and function

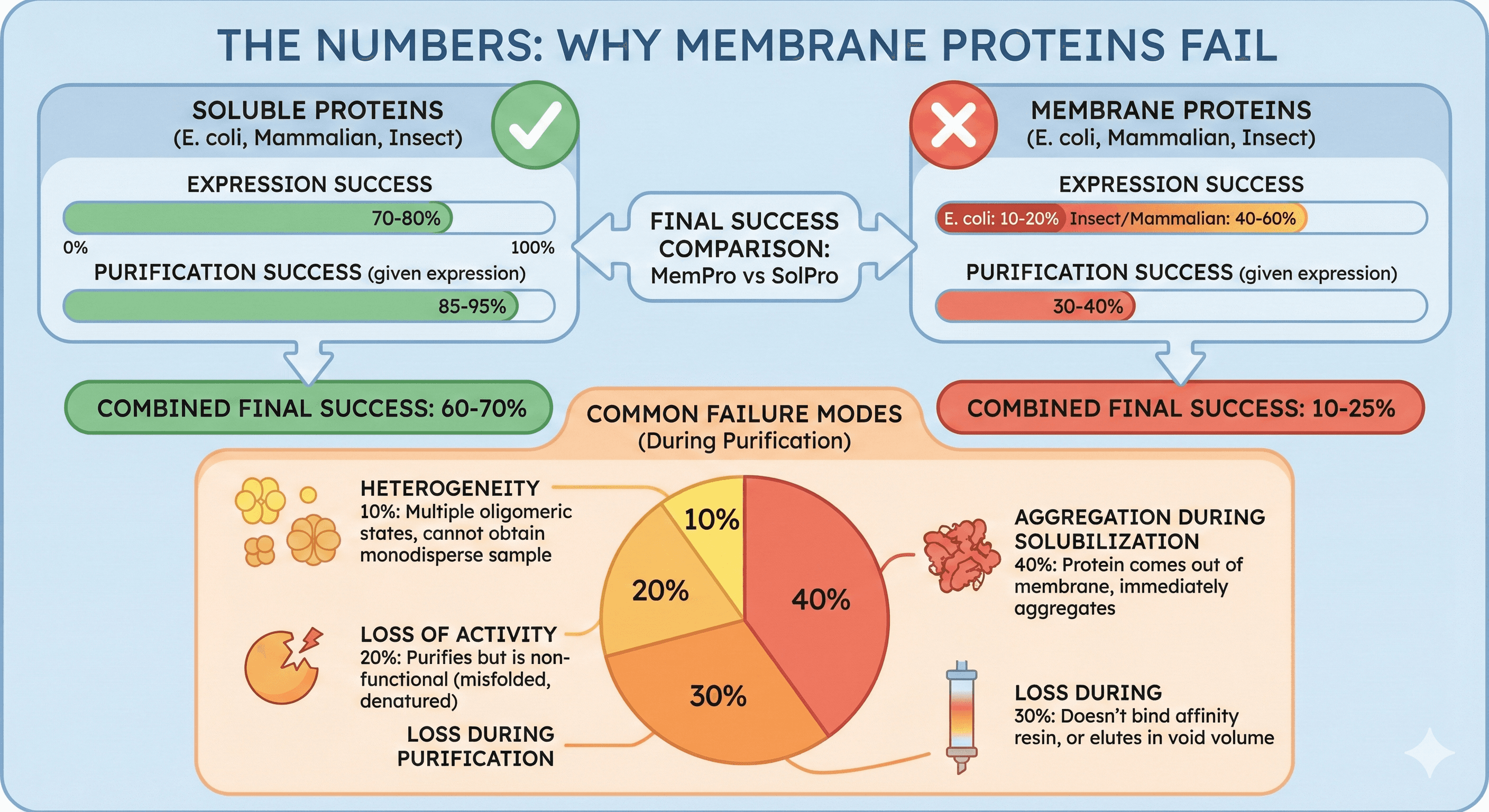
The Numbers: Why Membrane Proteins Fail
Expression success rate:
Soluble proteins (E. coli): 70-80%
Membrane proteins (E. coli): 10-20%
Membrane proteins (insect/mammalian): 40-60%
Purification success rate (given successful expression):
Soluble proteins: 85-95%
Membrane proteins: 30-40%
Combined (expression + purification):
Soluble proteins: 60-70% final success
Membrane proteins: 10-25% final success
Common failure modes:
Aggregation during solubilization (40%): Protein comes out of membrane, immediately aggregates
Loss during purification (30%): Doesn't bind affinity resin, or elutes in void volume
Loss of activity (20%): Purifies but is non-functional (misfolded, denatured)
Heterogeneity (10%): Multiple oligomeric states, cannot obtain monodisperse sample
Membrane Protein Classes
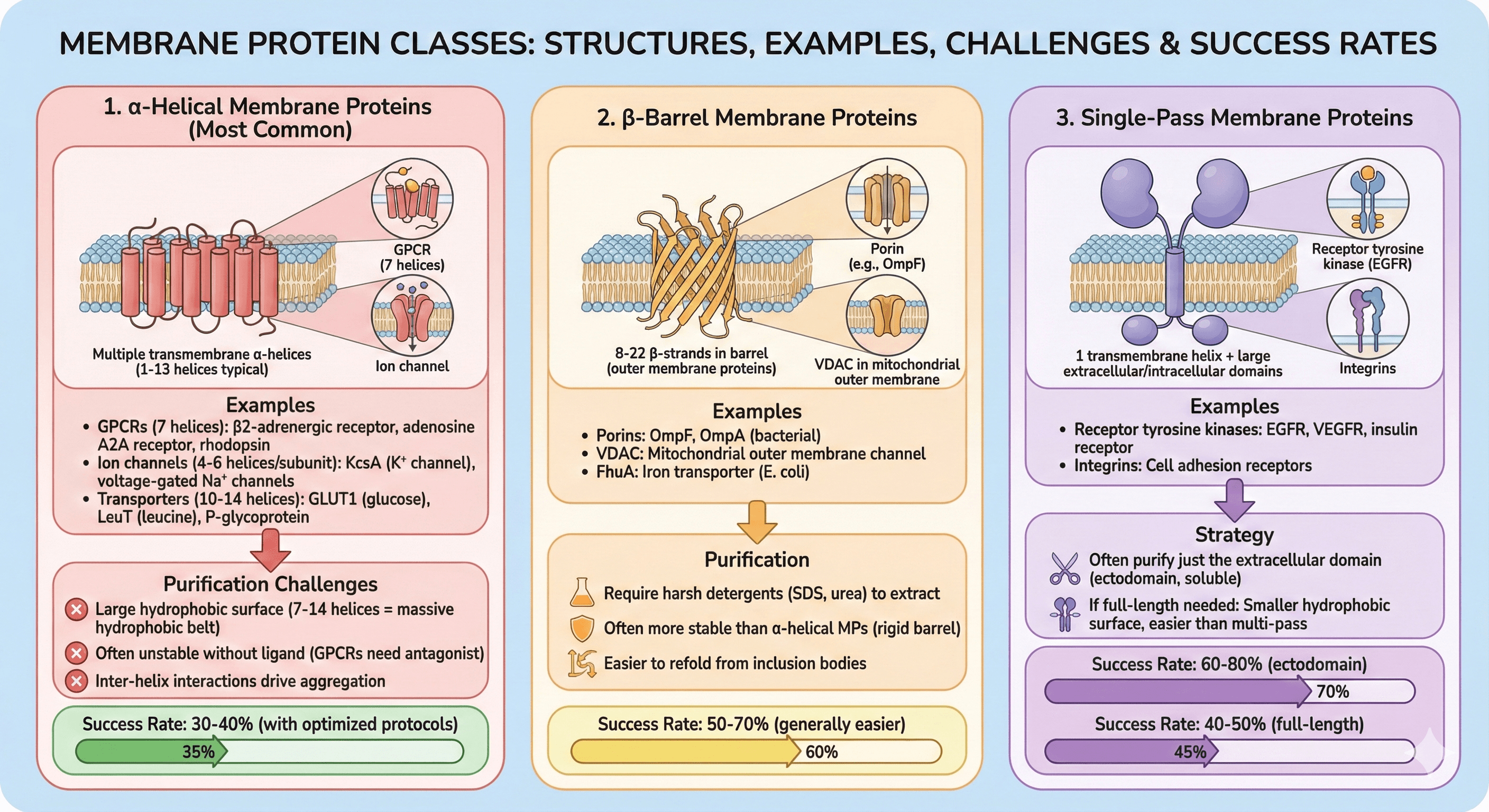
1. α-Helical Membrane Proteins (Most Common)
Structure: Multiple transmembrane α-helices (1-13 helices typical)
Examples:
GPCRs (7 helices): β2-adrenergic receptor, adenosine A2A receptor, rhodopsin
Ion channels (4-6 helices per subunit): KcsA (K⁺ channel), voltage-gated Na⁺ channels
Transporters (10-14 helices): GLUT1 (glucose), LeuT (leucine), P-glycoprotein
Purification challenges:
Large hydrophobic surface (7-14 helices = massive hydrophobic belt)
Often unstable without ligand (GPCRs need antagonist)
Inter-helix interactions drive aggregation
Success rate: 30-40% (with optimized protocols)
2. β-Barrel Membrane Proteins
Structure: 8-22 β-strands in barrel (outer membrane proteins)
Examples:
Porins: OmpF, OmpA (bacterial)
VDAC: Mitochondrial outer membrane channel
FhuA: Iron transporter (E. coli)
Purification:
Require harsh detergents (SDS, urea) to extract
Often more stable than α-helical MPs (rigid barrel)
Easier to refold from inclusion bodies
Success rate: 50-70% (generally easier)
3. Single-Pass Membrane Proteins
Structure: 1 transmembrane helix + large extracellular/intracellular domains
Examples:
Receptor tyrosine kinases: EGFR, VEGFR, insulin receptor
Integrins: Cell adhesion receptors
Strategy:
Often purify just the extracellular domain (ectodomain, soluble)
If full-length needed: Smaller hydrophobic surface, easier than multi-pass
Success rate: 60-80% (ectodomain), 40-50% (full-length)
The Purification Workflow: Stage-by-Stage
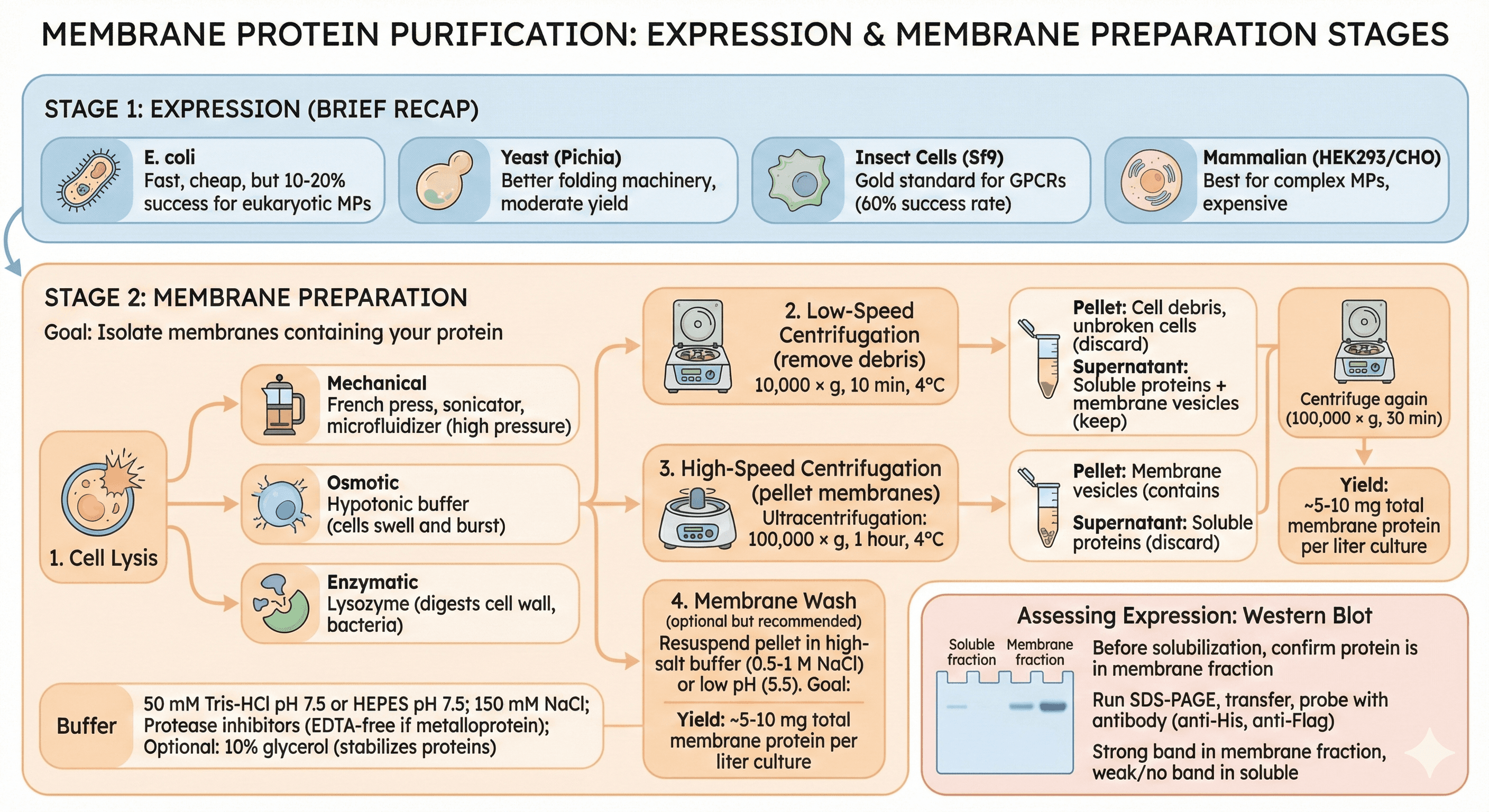
Stage 1: Expression (Brief Recap)
Expression system selection:
E. coli: Fast, cheap, but 10-20% success for eukaryotic MPs
Yeast (Pichia): Better folding machinery, moderate yield
Insect cells (Sf9): Gold standard for GPCRs (60% success rate)
Mammalian (HEK293/CHO): Best for complex MPs, expensive
Stage 2: Membrane Preparation
Goal: Isolate membranes containing your protein
Protocol:
1. Cell Lysis
Mechanical: French press, sonicator, microfluidizer (high pressure)
Osmotic: Hypotonic buffer (cells swell and burst)
Enzymatic: Lysozyme (digests cell wall, bacteria)
Buffer:
50 mM Tris-HCl pH 7.5 or HEPES pH 7.5
150 mM NaCl
Protease inhibitors (EDTA-free if metalloprotein)
Optional: 10% glycerol (stabilizes proteins)
2. Low-Speed Centrifugation (remove debris)
10,000 × g, 10 min, 4°C
Pellet: Cell debris, unbroken cells (discard)
Supernatant: Soluble proteins + membrane vesicles (keep)
3. High-Speed Centrifugation (pellet membranes)
Ultracentrifugation: 100,000 × g, 1 hour, 4°C
Pellet: Membrane vesicles (contains your MP)
Supernatant: Soluble proteins (discard)
4. Membrane Wash (optional but recommended)
Resuspend pellet in high-salt buffer (0.5-1 M NaCl) or low pH (5.5)
Goal: Remove peripherally associated proteins
Centrifuge again (100,000 × g, 30 min)
Yield: ~5-10 mg total membrane protein per liter culture
Assessing Expression: Western Blot
Before solubilization, confirm protein is in membrane fraction
Sample: Soluble fraction + membrane fraction
Run SDS-PAGE, transfer, probe with antibody (anti-His, anti-Flag)
Expected: Strong band in membrane fraction, weak/no band in soluble
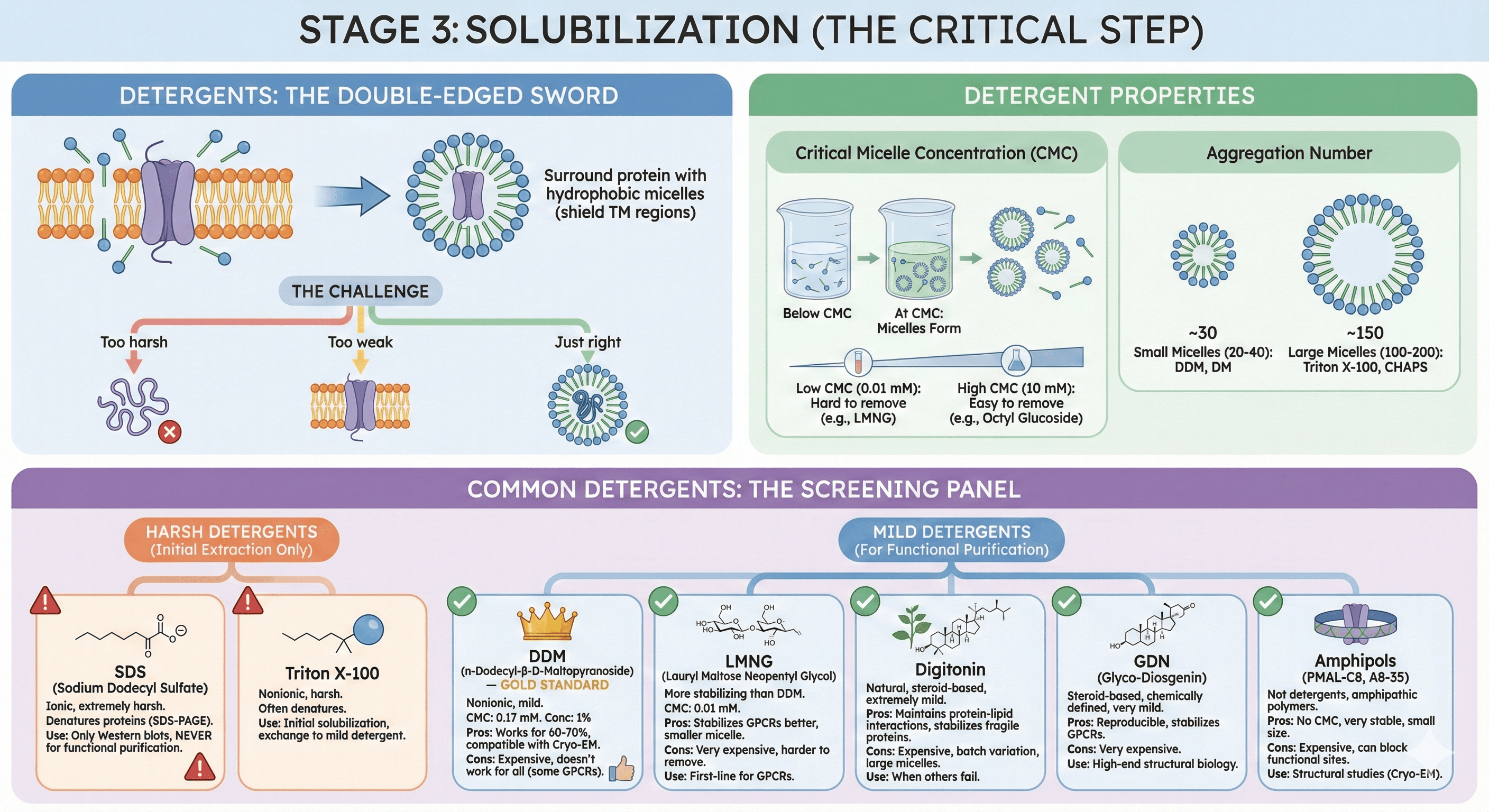
Stage 3: Solubilization (The Critical Step)
Detergents: The Double-Edged Sword
What detergents do:
Disrupt lipid bilayer (insert into membrane)
Surround protein with hydrophobic micelles (shield TM regions)
Allow protein to exist in aqueous solution
The challenge:
Too harsh: Protein denatures (unfolds, loses structure)
Too weak: Protein doesn't solubilize (stays in membrane)
Just right: Protein solubilizes, stays folded and functional
Detergent Properties
Critical Micelle Concentration (CMC):
Concentration at which detergent forms micelles
Low CMC (0.01 mM): Hard to remove (stays bound), e.g., LMNG
High CMC (10 mM): Easy to remove (dialyzes out), e.g., octyl glucoside
Aggregation number:
Number of detergent molecules per micelle
Small micelles (20-40): DDM, DM
Large micelles (100-200): Triton X-100, CHAPS
Common Detergents: The Screening Panel
Harsh Detergents (Initial Extraction Only)
1. SDS (Sodium Dodecyl Sulfate)
Ionic, extremely harsh
Denatures proteins (used in SDS-PAGE)
Use: Only for Western blots, never for functional purification
2. Triton X-100
Nonionic, harsh
Often denatures membrane proteins
Use: Sometimes for initial solubilization, but exchange to milder detergent
Mild Detergents (For Functional Purification)
3. DDM (n-Dodecyl-β-D-Maltopyranoside) — GOLD STANDARD
Nonionic, mild, stabilizes many MPs
CMC: 0.17 mM (relatively easy to remove)
Concentration: 1% (w/v) for solubilization, 0.02-0.05% for purification
Pros:
Works for 60-70% of membrane proteins
Well-tolerated
Compatible with crystallization, Cryo-EM
Cons:
Doesn't work for all proteins (some GPCRs aggregate)
Expensive (~$100-200/g)
4. LMNG (Lauryl Maltose Neopentyl Glycol)
Maltose-based, modified linker
More stabilizing than DDM (better for challenging targets)
CMC: 0.01 mM (harder to remove)
Pros:
Stabilizes GPCRs better than DDM (β2-AR, A2A receptor)
Smaller micelle size (better for Cryo-EM)
Cons:
More expensive (~5-10× DDM cost)
Harder to remove (low CMC)
Use: First-line for GPCRs
5. Digitonin
Natural product (from Digitalis plants)
Steroid-based, extremely mild
Preserves native lipids around protein
Pros:
Maintains protein-lipid interactions
Stabilizes very fragile proteins (respiratory complexes, some GPCRs)
Cons:
Expensive ($50-100/g)
Heterogeneous (batch variation)
Large micelles (harder to crystallize)
Use: When everything else fails
6. GDN (Glyco-Diosgenin)
Steroid-based, like digitonin but chemically defined
Very mild, homogeneous
Pros:
Reproducible purifications
Stabilizes GPCRs (A2A receptor structure solved in GDN)
Cons:
Very expensive ($300-500/g)
Use: High-end structural biology
7. Amphipols (PMAL-C8, A8-35)
Not detergents, but amphipathic polymers
Wrap around protein like a belt
Pros:
No CMC (don't form micelles)
Very stable (proteins soluble for months)
Small size (better for Cryo-EM resolution)
Cons:
Expensive ($100-200/g)
Can block functional sites
Use: Structural studies (Cryo-EM), detergent-free environment
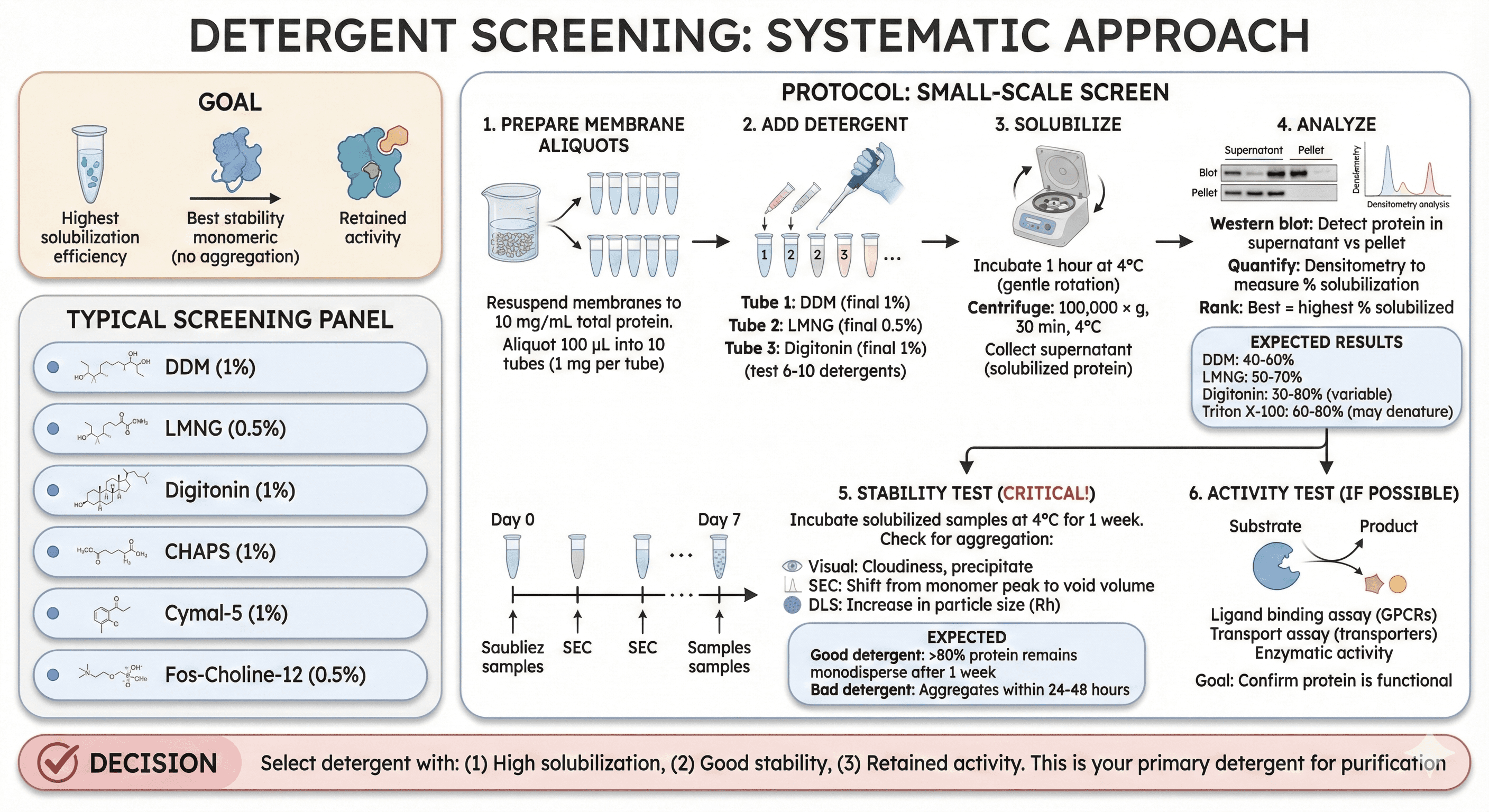
Detergent Screening: Systematic Approach
Goal: Find detergent that gives:
Highest solubilization efficiency
Best stability (no aggregation)
Retained activity
Typical screening panel:
DDM (1%)
LMNG (0.5%)
Digitonin (1%)
CHAPS (1%)
Cymal-5 (1%)
Fos-Choline-12 (0.5%)
Protocol: Small-Scale Screen
1. Prepare Membrane Aliquots
Resuspend membranes to 10 mg/mL total protein
Aliquot 100 μL into 10 tubes (1 mg per tube)
2. Add Detergent
Tube 1: DDM (final 1%)
Tube 2: LMNG (final 0.5%)
Tube 3: Digitonin (final 1%)
... (test 6-10 detergents)
3. Solubilize
Incubate 1 hour at 4°C (gentle rotation)
Centrifuge: 100,000 × g, 30 min, 4°C
Collect supernatant (solubilized protein)
4. Analyze
Western blot: Detect protein in supernatant vs pellet
Quantify: Densitometry to measure % solubilization
Rank: Best = highest % solubilized
Expected results:
DDM: 40-60% solubilization
LMNG: 50-70%
Digitonin: 30-80% (variable)
Triton X-100: 60-80% (but may denature)
5. Stability Test (Critical!)
Incubate solubilized samples at 4°C for 1 week
Check for aggregation:
Visual: Cloudiness, precipitate
SEC: Shift from monomer peak to void volume
DLS: Increase in particle size (Rh)
Expected:
Good detergent: >80% protein remains monodisperse after 1 week
Bad detergent: Aggregates within 24-48 hours
6. Activity Test (If Possible)
Ligand binding assay (GPCRs)
Transport assay (transporters)
Enzymatic activity
Goal: Confirm protein is functional
Decision:
Select detergent with: (1) High solubilization, (2) Good stability, (3) Retained activity
This is your primary detergent for purification
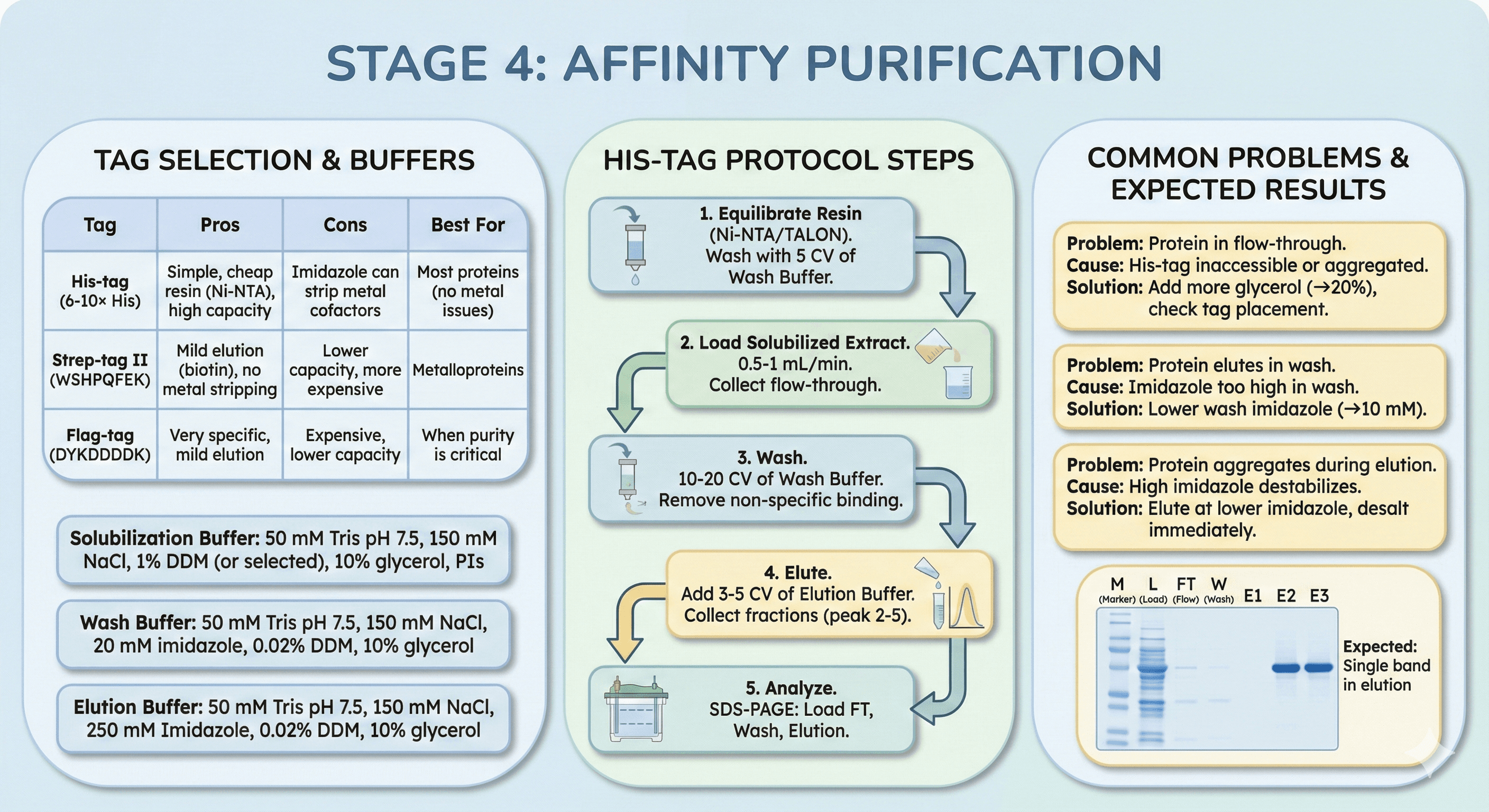
Stage 4: Affinity Purification
Tag Selection
His-tag (6-10× His):
Pros: Simple, cheap resin (Ni-NTA), high capacity
Cons: Imidazole can strip metal cofactors
Best for: Most proteins (no metal cofactor issues)
Strep-tag II (WSHPQFEK):
Pros: Mild elution (biotin), no metal stripping
Cons: Lower capacity, more expensive
Best for: Metalloproteins
Flag-tag (DYKDDDDK):
Pros: Very specific, mild elution
Cons: Expensive, lower capacity
Best for: When purity is critical
Protocol: His-Tag Affinity Purification
Buffers:
Solubilization buffer:
50 mM Tris pH 7.5, 150 mM NaCl
1% DDM (or selected detergent)
10% glycerol, protease inhibitors
Wash buffer:
50 mM Tris pH 7.5, 150 mM NaCl
20 mM imidazole (remove non-specific binding)
0.02% DDM (maintain solubilization, 10× CMC)
10% glycerol
Elution buffer:
50 mM Tris pH 7.5, 150 mM NaCl
250 mM imidazole
0.02% DDM
10% glycerol
Steps:
1. Equilibrate Resin
Ni-NTA or TALON (2-5 mL per L culture)
Wash with 5 CV of wash buffer
2. Load Solubilized Extract
Flow rate: 0.5-1 mL/min (gravity or pump)
Collect flow-through (save for analysis)
3. Wash
10-20 CV of wash buffer
Remove non-specific binding
4. Elute
Add 3-5 CV of elution buffer
Collect fractions (0.5-1 mL each)
Peak fractions: Typically fractions 2-5
5. Analyze
SDS-PAGE: Load flow-through, wash, elution
Expected: Single band at expected MW in elution fractions
Common Problems:
Problem 1: Protein in flow-through (doesn't bind)
Cause: His-tag inaccessible or protein aggregated
Solution: Add more glycerol (10% → 20%), check tag placement
Problem 2: Protein elutes in wash
Cause: Imidazole too high in wash
Solution: Lower wash imidazole (20 mM → 10 mM)
Problem 3: Protein aggregates during elution
Cause: High imidazole destabilizes
Solution: Elute at lower imidazole (150-250 mM), desalt immediately
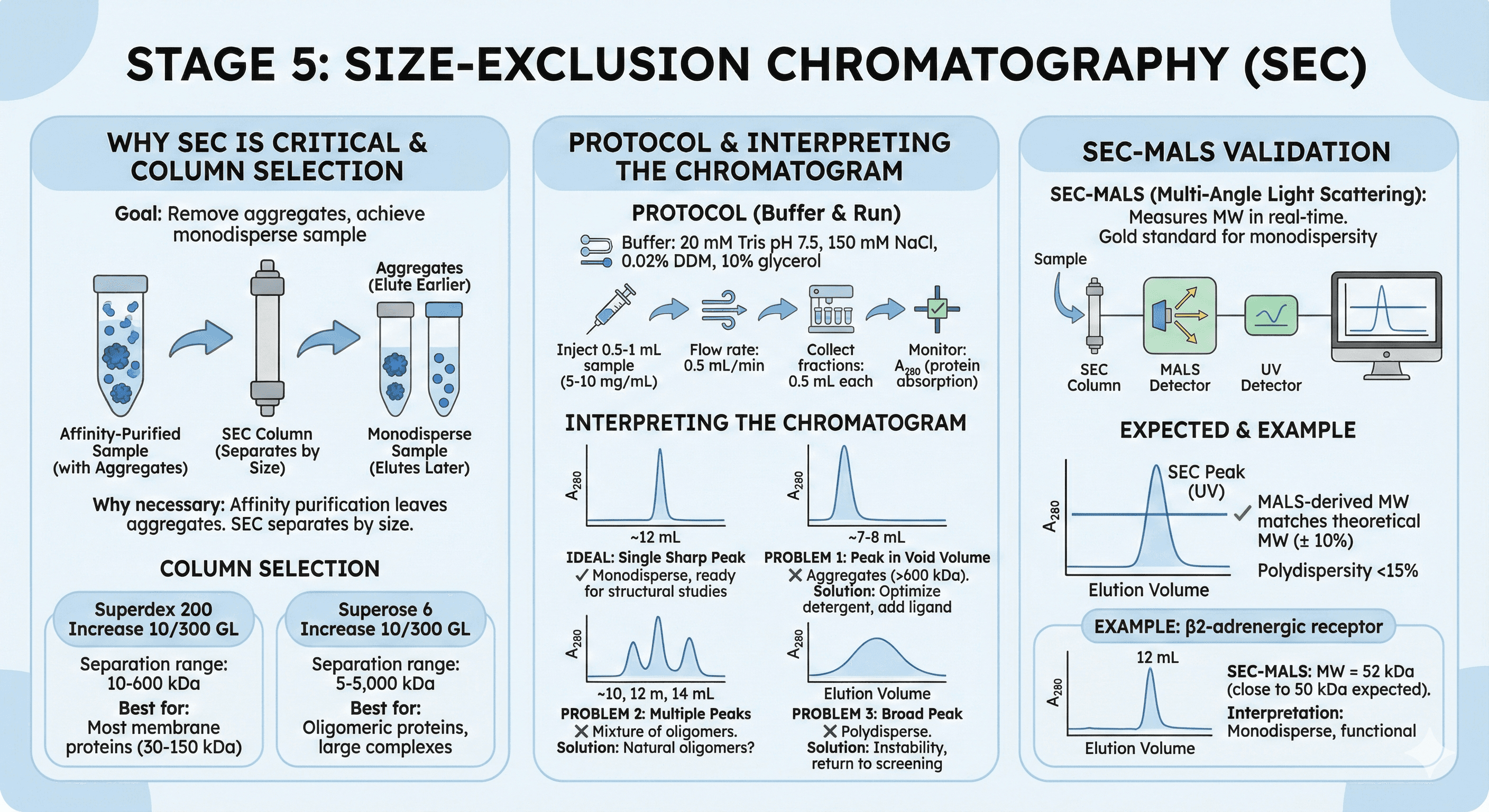
Stage 5: Size-Exclusion Chromatography (SEC)
Why SEC Is Critical
Goal: Remove aggregates, achieve monodisperse sample
Why necessary:
Affinity purification removes contaminants, but leaves aggregates
Aggregates interfere with structural studies (heterogeneous)
SEC separates by size: Monomers elute later, aggregates elute earlier
Column Selection
Superdex 200 Increase 10/300 GL:
Separation range: 10-600 kDa
Best for: Most membrane proteins (30-150 kDa)
Superose 6 Increase 10/300 GL:
Separation range: 5-5,000 kDa
Best for: Oligomeric proteins, large complexes
Protocol
Buffer:
20 mM Tris pH 7.5, 150 mM NaCl
0.02% DDM (or your detergent, 10× CMC)
10% glycerol
Run:
Inject 0.5-1 mL sample (5-10 mg/mL)
Flow rate: 0.5 mL/min
Collect fractions: 0.5 mL each
Monitor: A₂₈₀ (protein absorption)
Interpreting the Chromatogram
Ideal: Single sharp peak
Elution volume: 10-14 mL (for 30-50 kDa on Superdex 200)
Peak width: Narrow (Gaussian)
Interpretation: Monodisperse, ready for structural studies
Problem 1: Peak in void volume (7-8 mL)
Interpretation: Aggregates (>600 kDa)
Solution: Optimize detergent, add ligand, lower concentration
Problem 2: Multiple peaks
Interpretation: Mixture of oligomers (monomer, dimer, tetramer)
Solution: Some proteins are naturally oligomeric (e.g., ion channels)
Problem 3: Broad peak
Interpretation: Polydisperse (mixture of aggregation states)
Solution: Indicates instability, return to detergent screening
SEC-MALS (Validation)
SEC-MALS (Multi-Angle Light Scattering):
Measures molecular weight in real-time
Gold standard for confirming monodispersity
Expected:
SEC peak elutes at expected volume
MALS-derived MW matches theoretical MW (± 10%)
Polydispersity <15%
Example:
β2-adrenergic receptor (50 kDa with T4-lysozyme fusion)
SEC-MALS: MW = 52 kDa (close to expected)
Interpretation: Monodisperse, functional
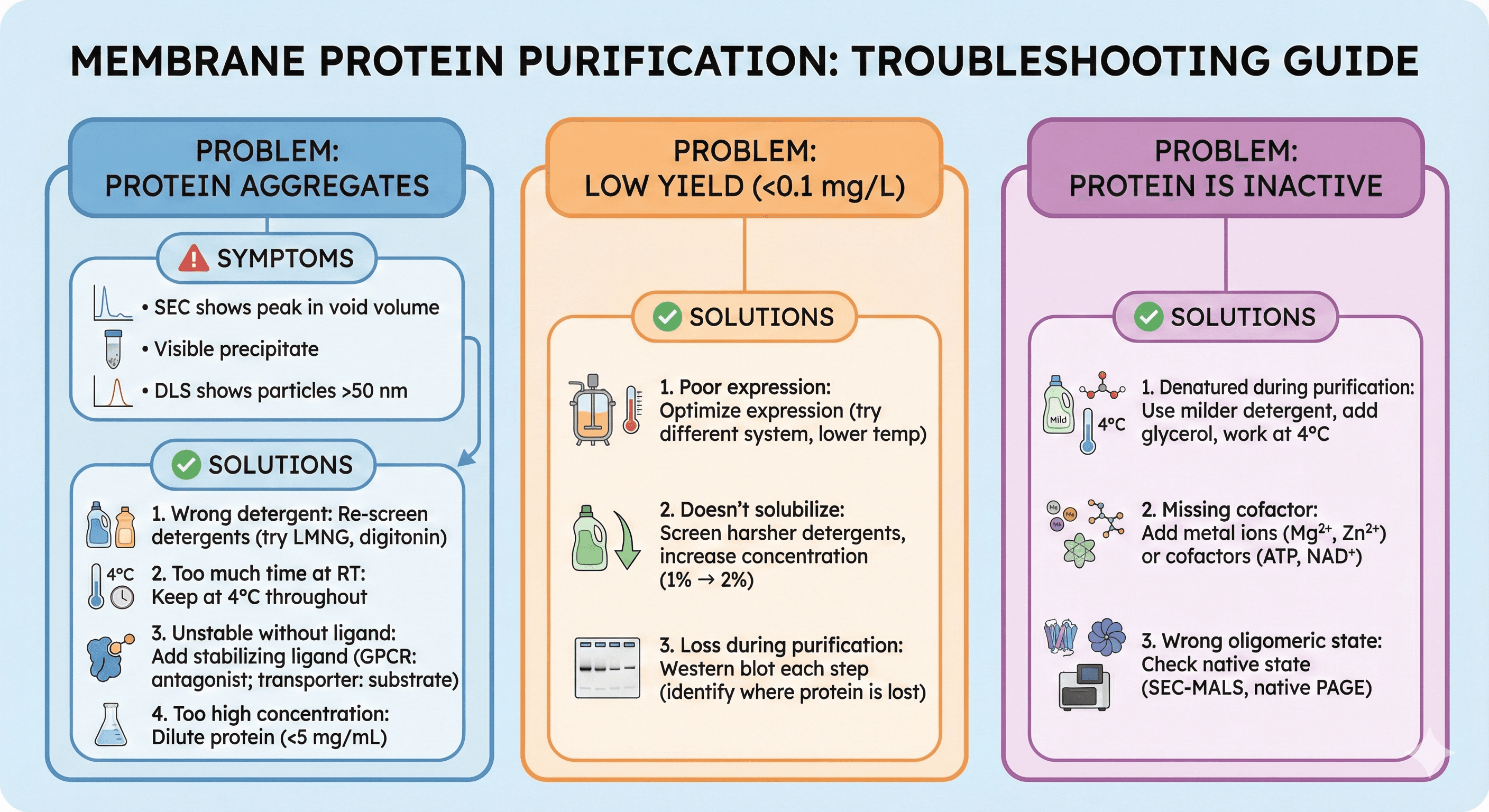
Troubleshooting Guide
Problem: Protein aggregates during purification
Symptoms:
SEC shows peak in void volume
Visible precipitate
DLS shows particles >50 nm
Solutions:
1. Wrong detergent
Re-screen detergents (try LMNG, digitonin)
2. Too much time at room temperature
Keep at 4°C throughout
3. Unstable without ligand
Add stabilizing ligand (GPCR: antagonist; transporter: substrate)
4. Too high concentration
Dilute protein (<5 mg/mL)
Problem: Low yield (<0.1 mg/L)
Solutions:
1. Poor expression
Optimize expression (try different system, lower temperature)
2. Doesn't solubilize
Screen harsher detergents, increase concentration (1% → 2%)
3. Loss during purification
Western blot each step (identify where protein is lost)
Problem: Protein is inactive
Solutions:
1. Denatured during purification
Use milder detergent, add glycerol, work at 4°C
2. Missing cofactor
Add metal ions (Mg²⁺, Zn²⁺) or cofactors (ATP, NAD⁺)
3. Wrong oligomeric state
Check native state (SEC-MALS, native PAGE)
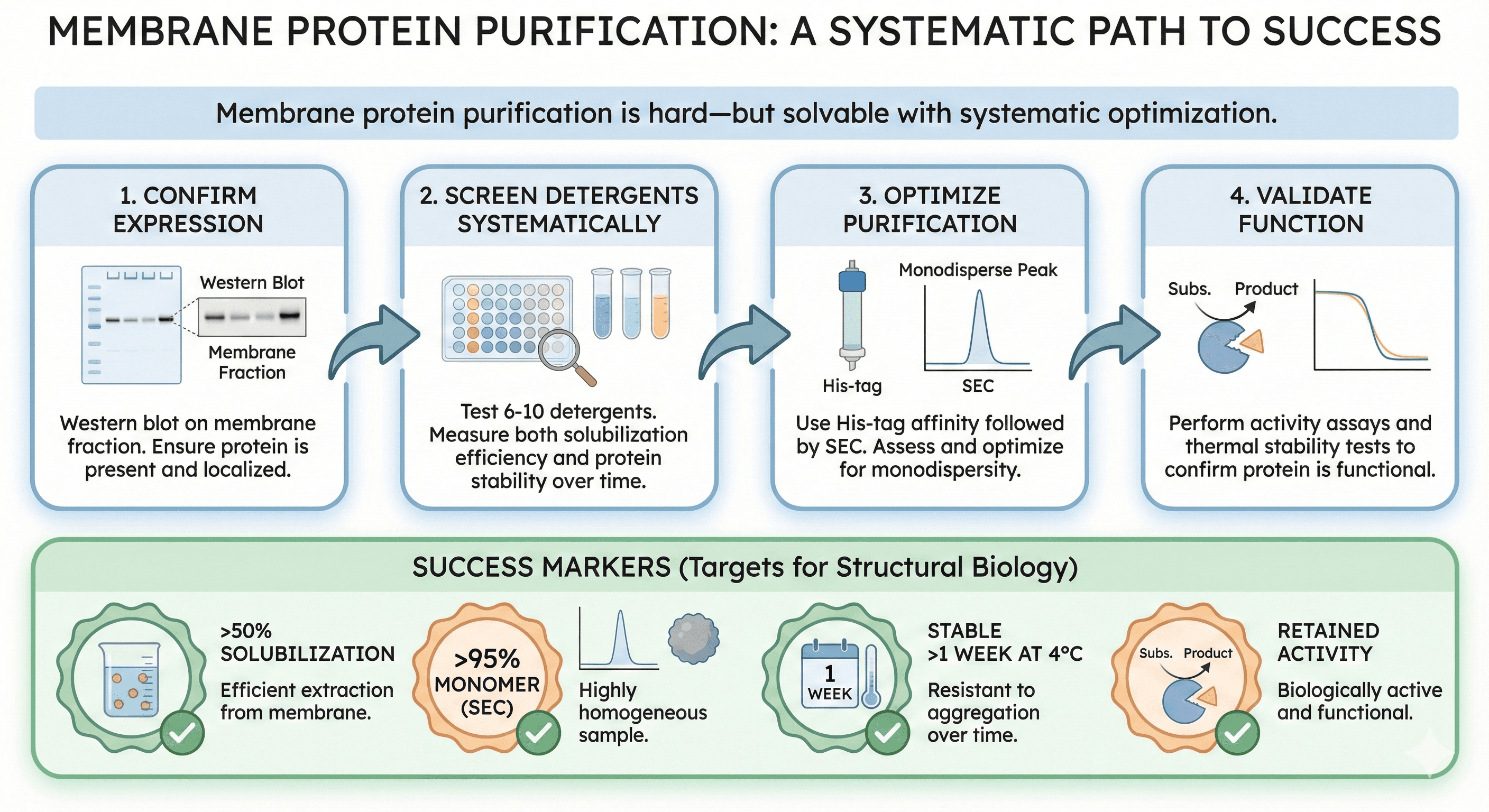
The Bottom Line
Membrane protein purification is hard—but solvable with systematic optimization.
The key steps:
Confirm expression (Western blot on membrane fraction)
Screen detergents systematically (test 6-10, measure solubilization + stability)
Optimize purification (His-tag, SEC, assess monodispersity)
Validate function (activity assay, thermal stability)
Success markers:
50% solubilization
95% monomer (SEC)
Stable >1 week at 4°C
Retained activity
Ready to Optimize Your Membrane Protein Purification?
If you're struggling with membrane protein stability or activity, Orbion can help design better constructs.
Orbion provides:
Thermostabilizing mutation predictions (increase Tm, reduce aggregation)
Construct boundary design (truncate disordered regions)
Membrane topology prediction (identify TM helices, loop regions)
Expression system recommendations
PTM prediction (glycosylation sites requiring mammalian expression)