
Updates
Product
Launch
Complex Predictions Now Live — Structure to Protocol in One Workflow
Dec 8, 2025
Most complex structure projects fail at construct design, not data collection.
You express multiple chains, get aggregation or substoichiometric assembly, and optimize blindly for months. Standard AlphaFold-Multimer gives structure but no insight into interface reliability, flexibility, or how to design working constructs.
Now available: Complex prediction and analysis on Orbion - from sequences to construct strategy in one workflow.
Platform Capabilities:
AlphaFold-Multimer + dual interface confidence (chain-to-chain + geometry)
Mechanistic roles (receptor/ligand, scaffold/regulator/connector)
Flexibility mapping (truncation candidates, hinges, multi-state residues)
Functional predictions (PTMs, binding sites, membrane topology)
Integrated co-expression protocols (stoichiometry, mutations, additives)
Works for: Antibody-antigen, enzyme complexes, membrane assemblies, kinase-substrate, protein-peptide.

Example: GLP-1R–GLP-1 Complex
GPCR-peptide (1:1 hetero-oligomer). Clinically relevant (semaglutide mechanism), experimentally validated (PDB: 6X18, 5VAI), notoriously difficult (membrane protein + transient binding + DPP4 degradation + glycosylation interference).
Structure + Interface Predictions:
Dual confidence (chain 18.4Å, geometry 3.1Å) explains why binding interface is reliable despite conformational dynamics
4 flexibility zones (N-term, ICL2/3, C-term) ✅ match truncations in solved structures
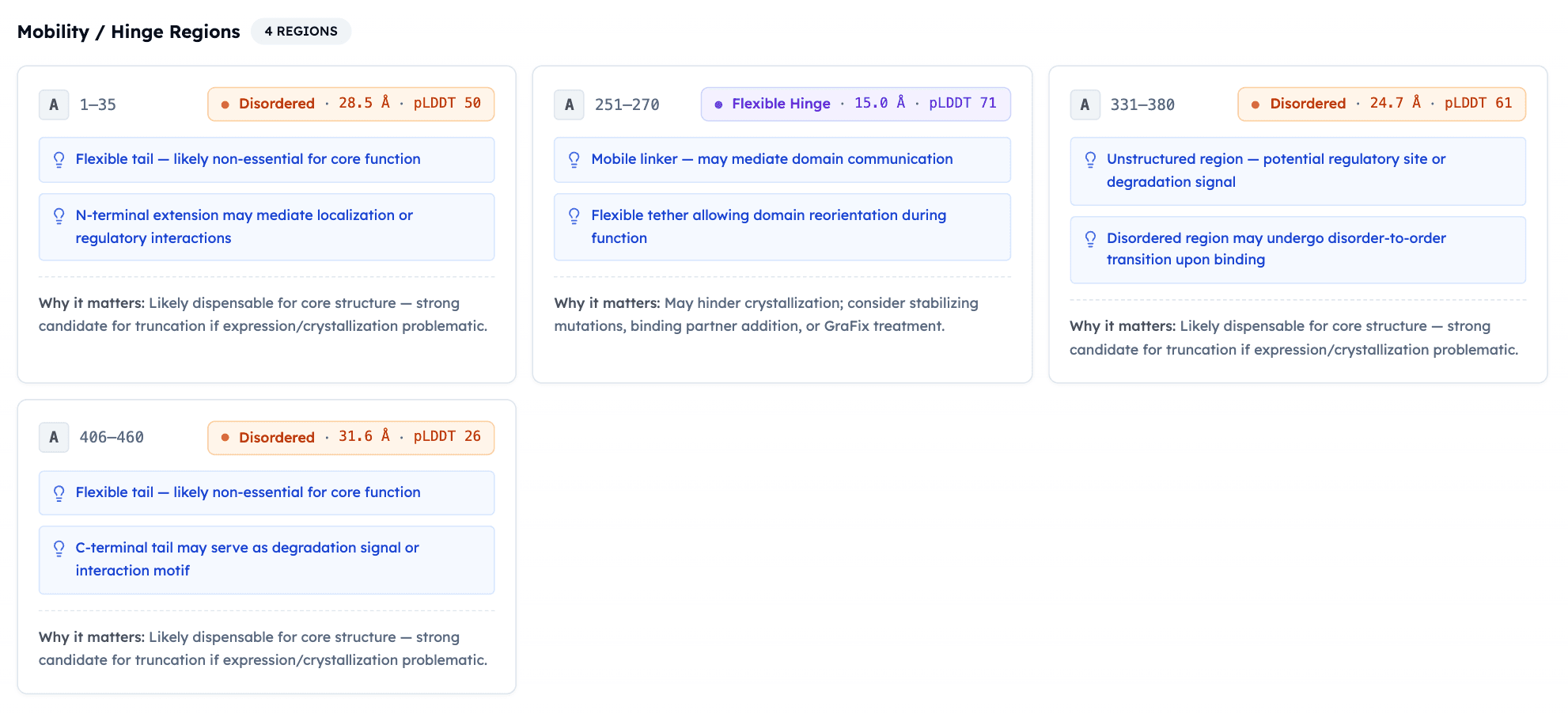
Functional Sites:
4 disulfide bridges (matches PDB ✅)
N-glycosylation (N63, N82) (expression-critical ✅)
Phosphorylation cluster (S431-444) in desensitization region
Cholesterol binding sites
GLP-1 C-term amidation (required for activity ✅)
Each structural prediction chains into experimental decisions:
N-glycosylation (N63, N82) → HEK293S GnTI⁻ at 31-32°C for glycan control
Cholesterol binding sites → LMNG/CHS detergent + POPC nanodisc composition
DPP4 cleavage site → Sitagliptin (10 μM) + A8G mutation for dual protection
Conformational dynamics (PAE 18.4Å) → Crosslinking/SAXS validation recommendation
Additional parameters: DNA ratio 1:2 + Sodium butyrate (3-5 mM, 16h) + ELISA checkpoint
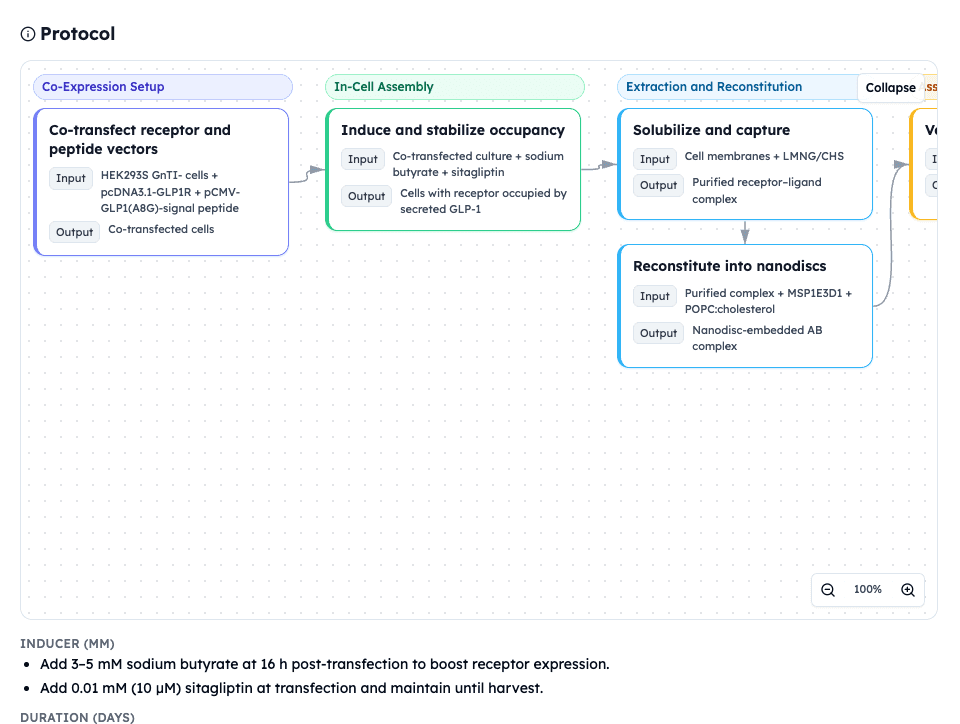
Result: Complete construct strategy with interconnected parameters that normally require multiple failed expression rounds. <30 minutes from sequences.

