Blog
Structure-Based Drug Design: From Protein Structure to Lead Compound
Dec 9, 2025
AlphaFold gave us the structure. Now what? With over 200 million predicted protein structures available, the bottleneck in drug discovery has shifted from "Can we get the structure?" to "Can we find druggable binding sites and design molecules that bind them?" Here's how structure-based drug design transforms a PDB file into a therapeutic candidate.
Key Takeaways
Structural information has contributed to the discovery or optimization of a large fraction of modern small-molecule drugs (~50% often quoted), especially in kinases, proteases, and antivirals
Main advantage: Rational design based on atomic-level interactions vs phenotypic screening
Success rate: Multiple studies report 2-5× improvements in hit-to-lead efficiency when SBDD is layered on top of HTS, rather than relying on HTS alone
Modern workflow: AlphaFold structure → Binding site prediction → Virtual screening → Lead optimization
Critical gap: AlphaFold predicts apo (ligand-free) states; drugs often require holo (ligand-bound) conformations
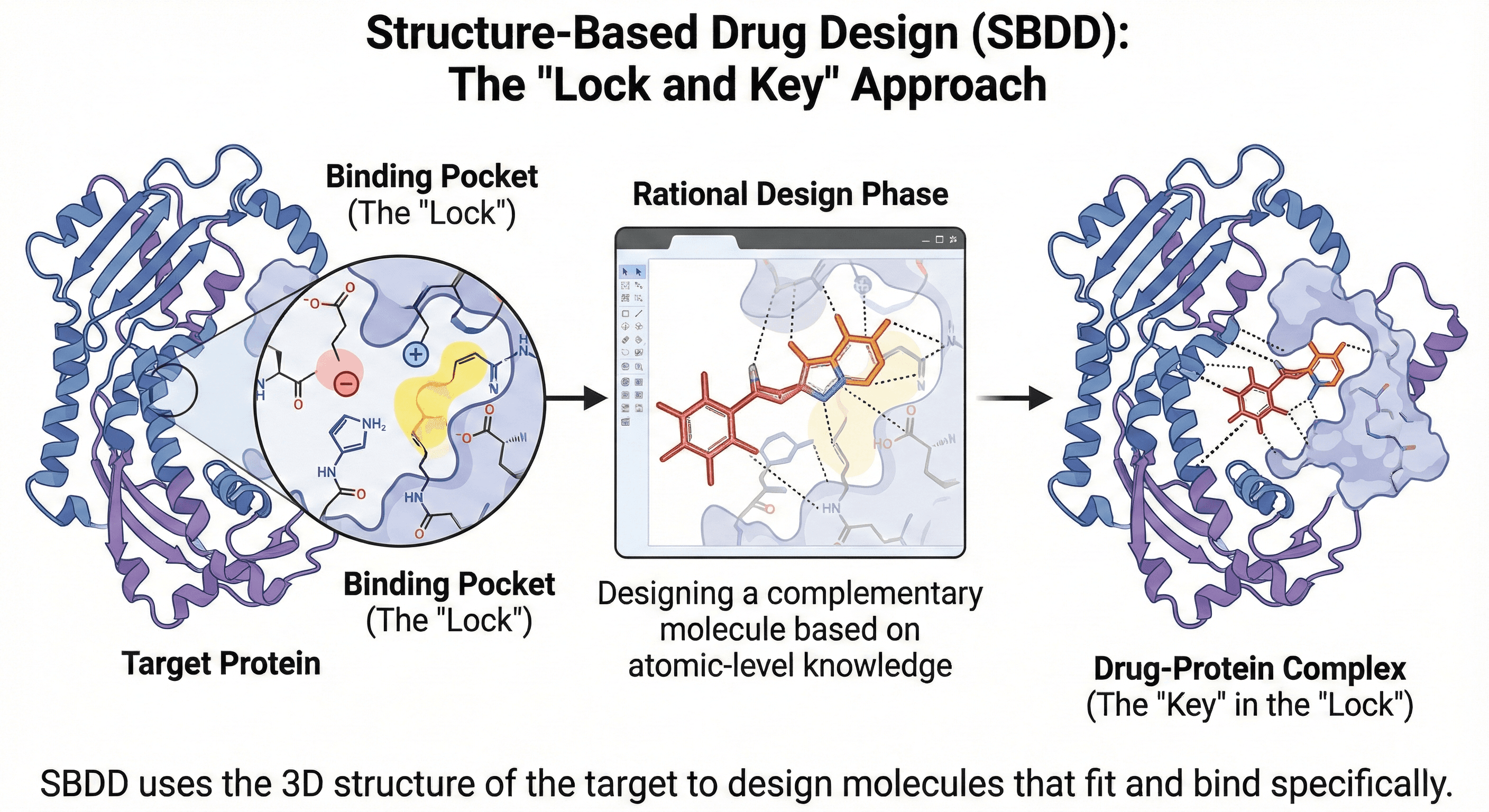
What Is Structure-Based Drug Design?
Structure-based drug design (SBDD) uses the three-dimensional structure of a protein target to guide the discovery and optimization of drug molecules. Instead of randomly testing millions of compounds hoping one sticks, we use atomic-level knowledge of the binding site to design molecules that should bind.
The concept is elegantly simple: If you know the shape and chemistry of a protein's binding pocket, you can design a molecule that fits like a key in a lock.
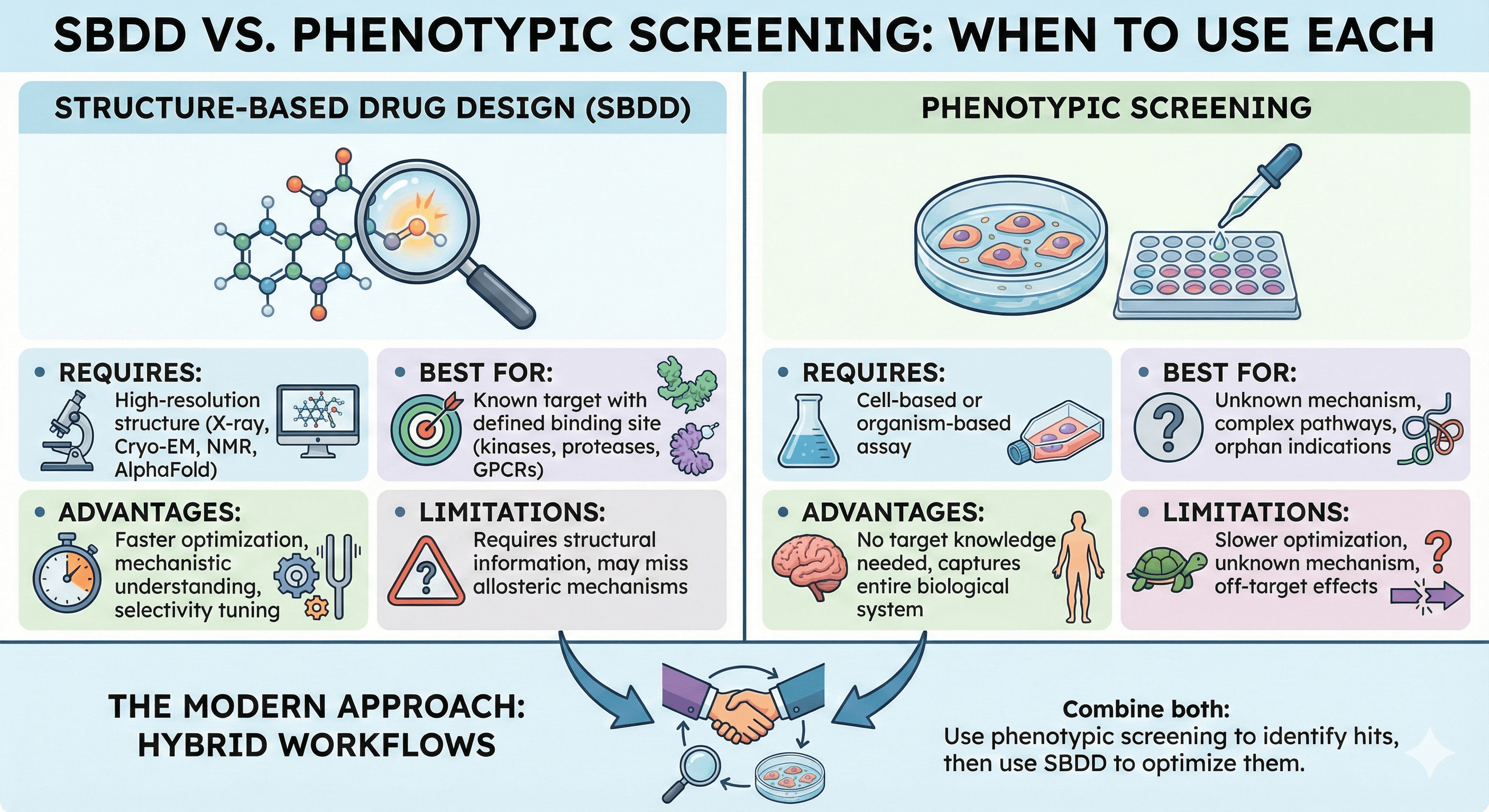
SBDD vs Phenotypic Screening: When to Use Each
Structure-Based Drug Design (SBDD):
Requires: High-resolution structure (X-ray, Cryo-EM, NMR, or AlphaFold)
Best for: Known target with defined binding site (kinases, proteases, GPCRs)
Advantages: Faster optimization, mechanistic understanding, selectivity tuning
Limitations: Requires structural information, may miss allosteric mechanisms
Phenotypic Screening:
Requires: Cell-based or organism-based assay
Best for: Unknown mechanism, complex pathways, orphan indications
Advantages: No target knowledge needed, captures entire biological system
Limitations: Slower optimization, unknown mechanism, off-target effects
The Modern Approach: Hybrid workflows that combine both—use phenotypic screening to identify hits, then use SBDD to optimize them.
The SBDD Workflow: Five Critical Steps
Quick Overview:
Get structure (X-ray, Cryo-EM, AlphaFold)
Find druggable sites (FPocket, Orbion)
Virtual screening (dock millions of compounds)
Optimize hits (improve potency, selectivity, ADMET)
Validate experimentally (biochemical + cell assays)
Below is the detailed breakdown of each step:
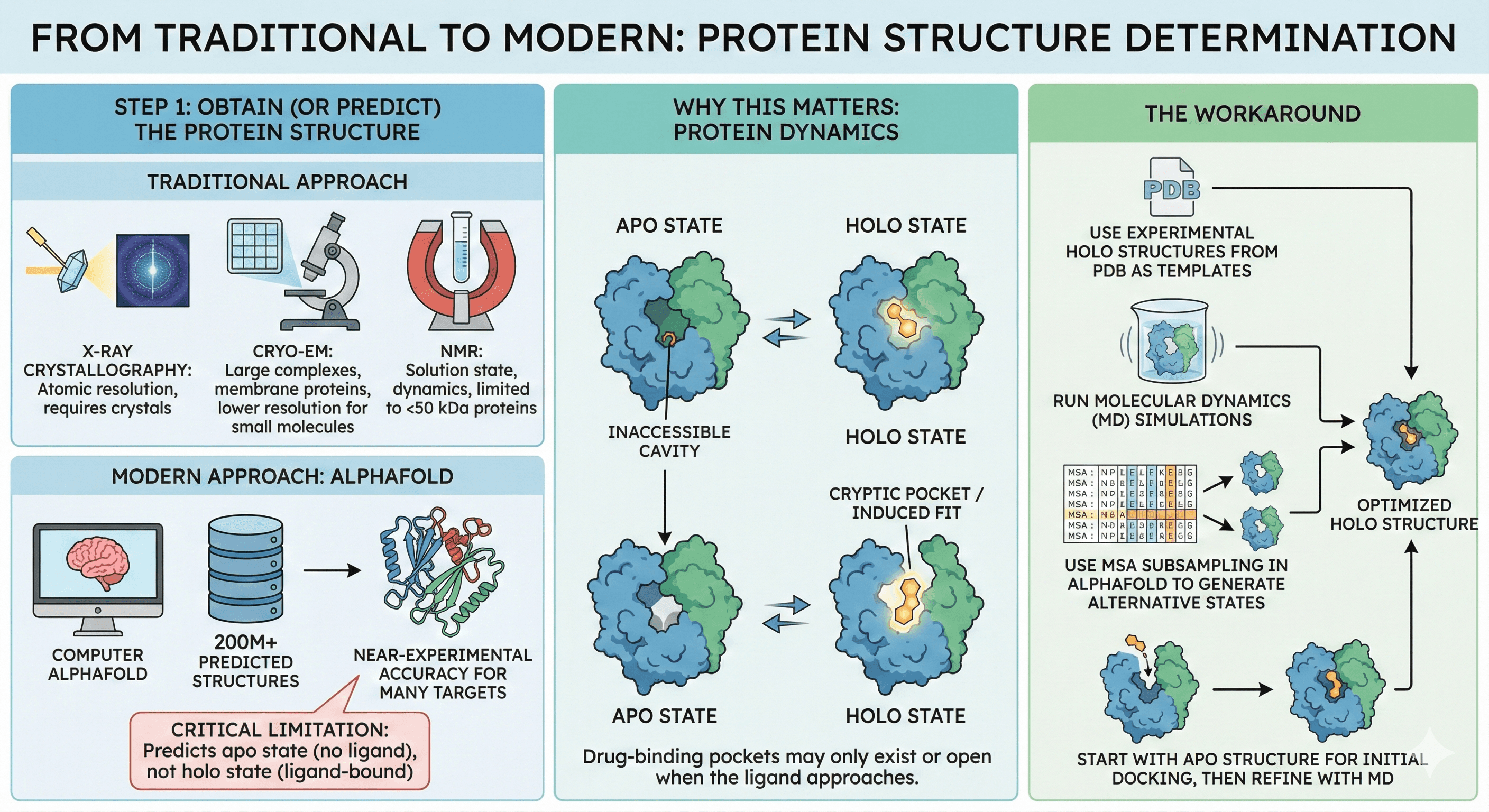
Step 1: Obtain (or Predict) the Protein Structure
Traditional approach:
X-ray crystallography (atomic resolution, but requires crystals)
Cryo-EM (large complexes, membrane proteins, but lower resolution for small molecules)
NMR (solution state, dynamics, but limited to <50 kDa proteins)
Modern approach: AlphaFold
200M+ predicted structures available
Near-experimental accuracy for many targets
Critical limitation: Predicts apo state (no ligand), not holo state (ligand-bound)
Why this matters: Proteins are dynamic. The pocket that binds a drug may not exist in the apo state—it might only open when the ligand approaches (cryptic pockets) or may adopt a different conformation when bound (induced fit).
The workaround:
Use experimental holo structures from PDB as templates
Run molecular dynamics (MD) simulations to sample conformations
Use MSA subsampling in AlphaFold to generate alternative states
Start with apo structure for initial docking, then refine with MD
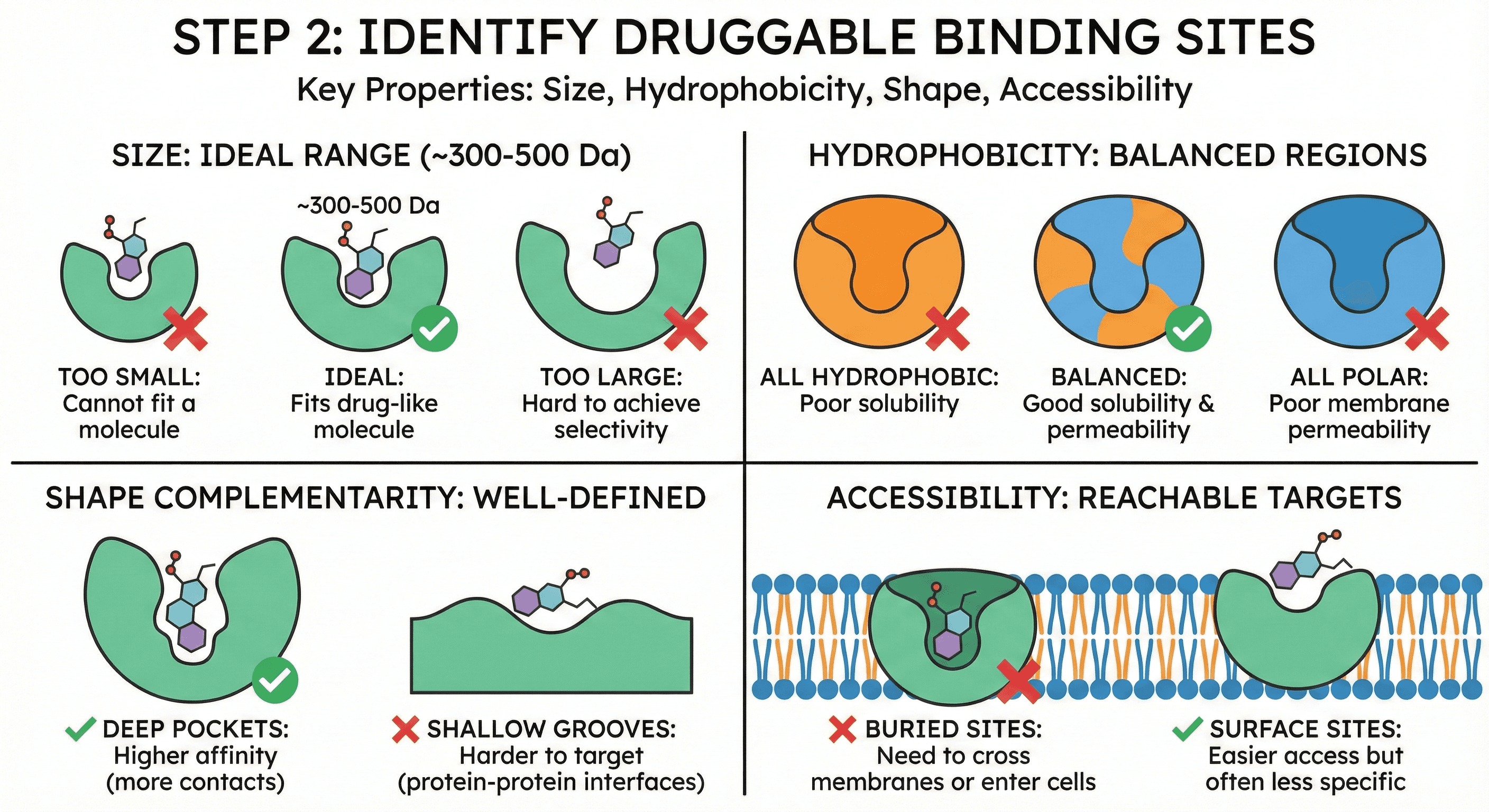
Step 2: Identify Druggable Binding Sites
Not all pockets are equal. A "druggable" binding site has specific properties:
Size: Large enough for a drug-like molecule (~300-500 Da)
Too small: Cannot fit a molecule
Too large: Hard to achieve selectivity
Hydrophobicity: Balance of hydrophobic and polar regions
All hydrophobic: Poor solubility
All polar: Poor membrane permeability
Shape complementarity: Well-defined pocket with clear boundaries
Deep pockets: Higher affinity (more contacts)
Shallow grooves: Harder to target (protein-protein interfaces)
Accessibility: Can a drug molecule reach the pocket?
Buried sites: Need to cross membranes or enter cells
Surface sites: Easier access but often less specific
Hot Sites vs Cold Sites
Hot sites (high druggability):
ATP-binding pockets in kinases
Protease active sites (HIV protease, NS3/4A in Hepatitis C)
GPCR orthosteric pockets
Cold sites (low druggability):
Flat protein-protein interfaces (historically "undruggable")
Large, solvent-exposed grooves
Sites with no clear pocket
Tools for Binding Site Prediction
FPocket / DoGSiteScorer: Geometry-based pocket detection SiteMap (Schrödinger): Druggability scoring P2Rank: Machine learning-based site prediction AlphaFill: Predicts ligand positions based on homology Orbion: Combines structure analysis with evolutionary conservation to identify functional binding sites
Example: Identifying cryptic pockets Cryptic pockets don't appear in static structures—they only open transiently. These are increasingly important drug targets because they often provide selectivity (not all homologs have the same cryptic pocket).
Case: BCL-2 family inhibitors
Static structure: No clear druggable pocket
Molecular dynamics: Transient pocket opens near BH3 binding groove
Result: Venetoclax (FDA-approved for CLL) targets this cryptic site
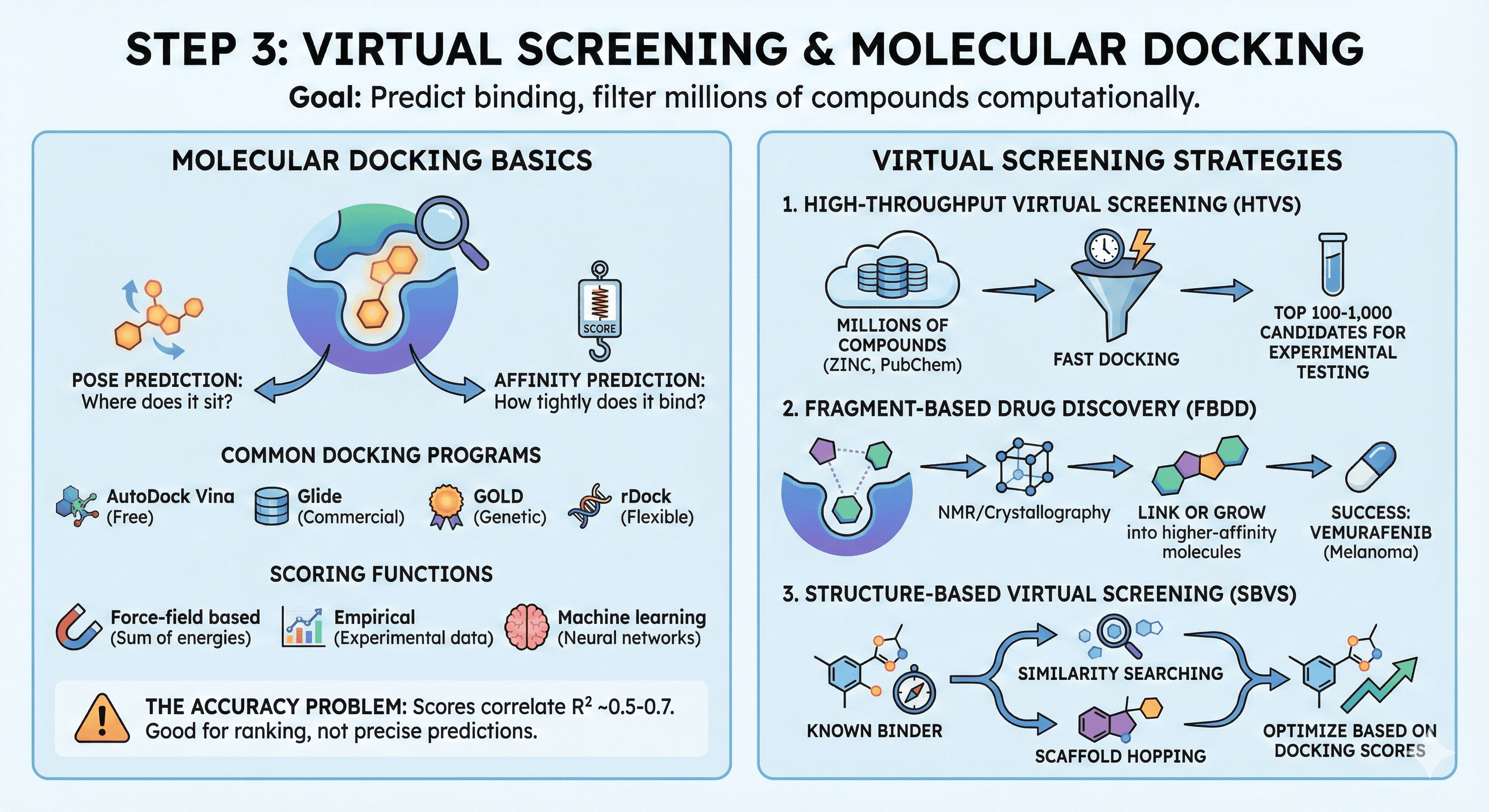
Step 3: Virtual Screening & Molecular Docking
Once you have a binding site, you need molecules that fit. Virtual screening tests millions of compounds computationally before synthesizing anything.
Molecular Docking Basics
The goal: Predict how a small molecule binds to the protein
Pose prediction: Where does the ligand sit?
Affinity prediction: How tightly does it bind?
Common docking programs:
AutoDock Vina (free, widely used)
Glide (Schrödinger) (commercial, high accuracy)
GOLD (genetic algorithm-based)
rDock (open-source, flexible)
Scoring functions: Docking programs estimate binding affinity using simplified energy functions:
Force-field based: Sum of van der Waals, electrostatics, H-bonds
Empirical: Trained on experimental binding data
Machine learning: Neural networks trained on protein-ligand complexes
The accuracy problem: Docking scores correlate with binding affinity with R² ~0.5-0.7. They're good for ranking candidates but not precise affinity predictions.
Virtual Screening Strategies
1. High-Throughput Virtual Screening (HTVS)
Screen 10⁶–10⁹ compounds from libraries (ZINC, PubChem, vendor catalogs)
Fast docking (seconds per compound)
Goal: Identify top 100-1,000 candidates for experimental testing
2. Fragment-Based Drug Discovery (FBDD)
Screen small fragments (~150 Da) with weak binding (mM range)
Crystallography or NMR to determine binding mode
Link or grow fragments into larger, higher-affinity molecules
Success: Vemurafenib (melanoma, targets BRAF V600E)
3. Structure-Based Virtual Screening (SBVS)
Use known binders as starting point
Similarity searching or scaffold hopping
Optimize based on docking scores
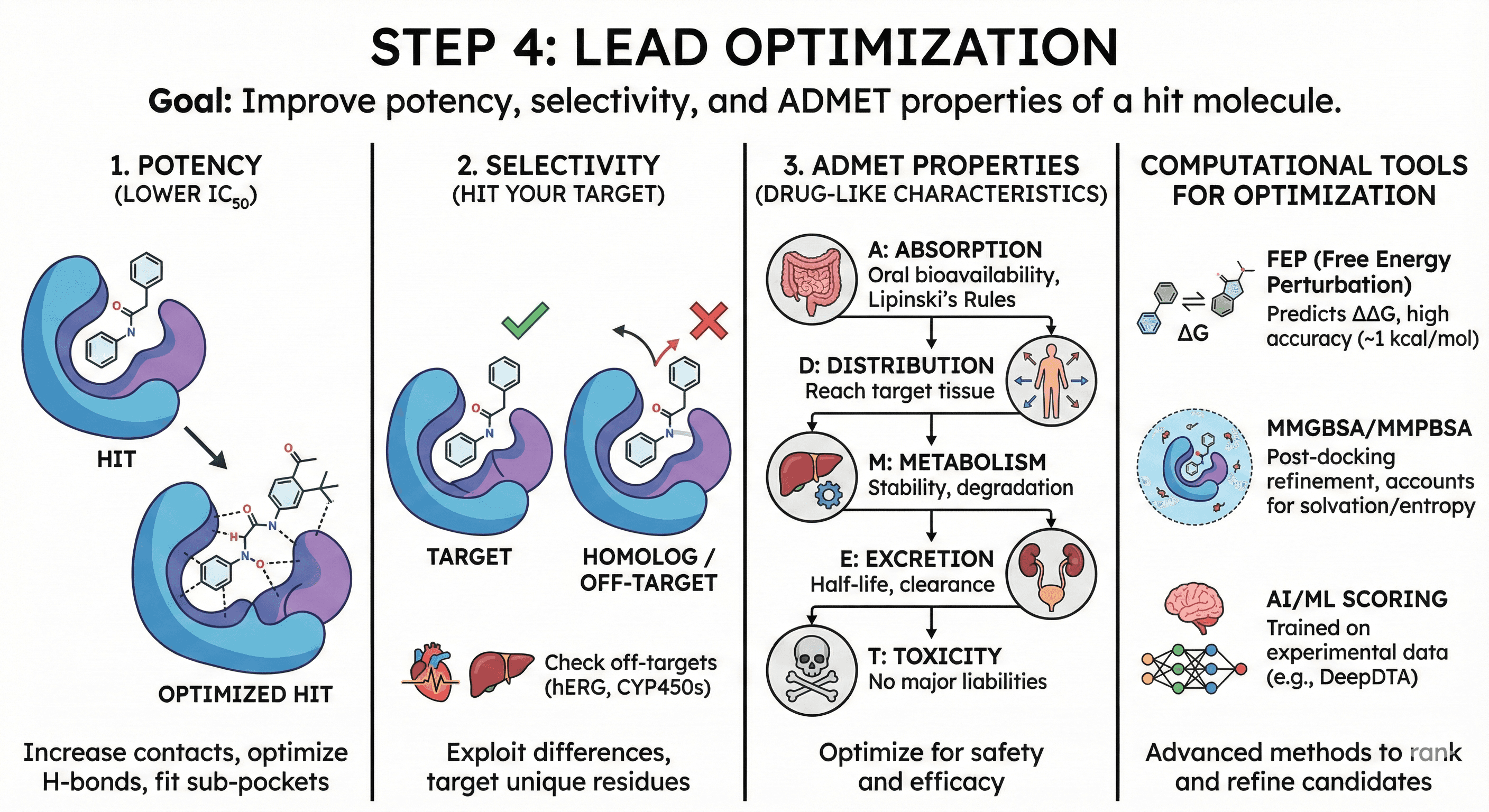
Step 4: Lead Optimization
You've found a hit (IC₅₀ ~10-100 μM). Now make it better.
The Three Pillars of Lead Optimization
1. Potency (lower IC₅₀)
Increase contacts with binding site
Optimize H-bonds, hydrophobic packing
Add functional groups that fit sub-pockets
2. Selectivity (hit your target, not others)
Exploit differences between homologs
Target unique residues in the binding site
Check off-target binding (hERG for cardiotoxicity, CYP450s for metabolism)
3. ADMET properties (drug-like characteristics)
Absorption: Oral bioavailability (Lipinski's Rule of Five)
Distribution: Can it reach the target tissue?
Metabolism: Is it stable or rapidly degraded?
Excretion: Half-life, clearance
Toxicity: No major liabilities
Computational Tools for Optimization
FEP (Free Energy Perturbation):
Predicts ΔΔG of binding for mutations/modifications
Accuracy: ~1 kcal/mol (good for ranking)
Example: Schrodinger FEP+ improved kinase inhibitor affinity 100×
MMGBSA/MMPBSA:
Post-docking energy refinement
Accounts for solvation, entropy
Faster than FEP, less accurate
AI/ML scoring:
DeepDTA, KDEEP, OnionNet
Trained on experimental binding data
Emerging as more accurate than classical scoring
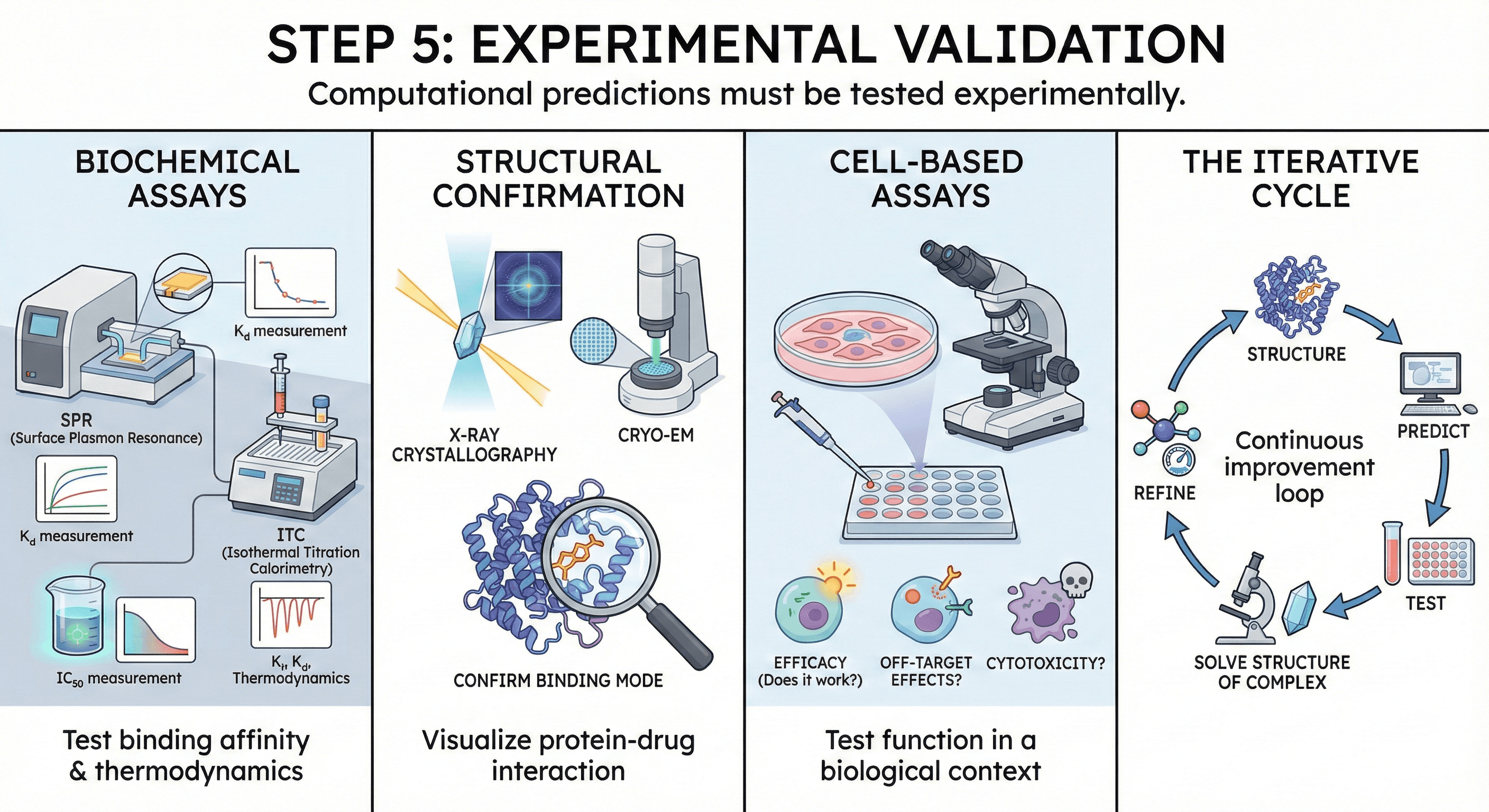
Step 5: Experimental Validation
Computational predictions must be tested experimentally.
Biochemical assays:
IC₅₀, Ki, Kd measurements
SPR (surface plasmon resonance), ITC (isothermal titration calorimetry)
Crystallography or Cryo-EM to confirm binding mode
Cell-based assays:
Does the compound work in cells?
Off-target effects?
Cytotoxicity?
Iterative cycle: Structure → Predict → Test → Solve structure of complex → Refine → Repeat
Case Study: HIV Protease Inhibitors
One of the greatest SBDD success stories.
The Target
HIV-1 protease: A homodimeric aspartic protease essential for viral replication. Without it, HIV produces non-infectious viral particles.
The Challenge
Small, symmetric active site
Highly flexible flaps cover the active site
Drug resistance mutations emerge quickly
The Solution (1990s SBDD)
Step 1: Crystal structure of HIV protease solved (1989)
Revealed symmetric active site with catalytic Asp25-Asp25' dyad
Identified substrate binding cleft
Step 2: Design peptidomimetic inhibitors
Mimic the natural peptide substrate
Replace scissile bond with non-cleavable transition state analog
Step 3: Optimize for drug-like properties
First-generation: Saquinavir (1995) - large, poor oral bioavailability
Second-generation: Ritonavir, Indinavir - improved pharmacokinetics
Third-generation: Darunavir (2006) - maintains activity against resistant mutants
Impact:
10+ FDA-approved HIV protease inhibitors
Part of HAART (highly active antiretroviral therapy)
Transformed HIV from death sentence to manageable chronic disease
Key lesson: Iterative structure-based optimization over 15+ years, combining X-ray crystallography of every new inhibitor with rational design, led to drugs that saved millions of lives.
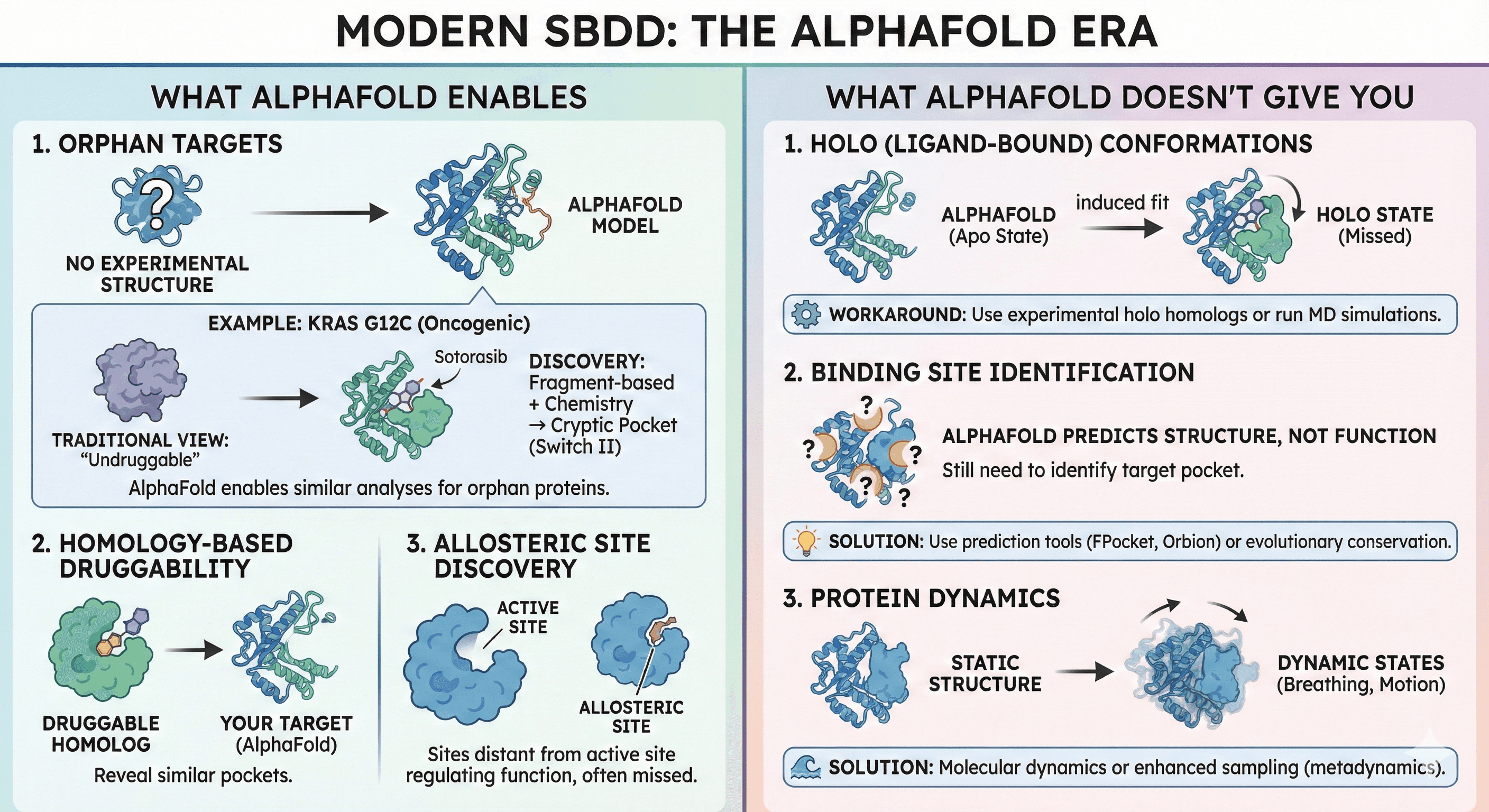
Modern SBDD: The AlphaFold Era
AlphaFold changes the game by making structures accessible for previously "unstructurable" targets.
What AlphaFold Enables
1. Orphan targets Proteins with no experimental structure can now be modeled with high confidence.
Example: KRAS G12C (oncogenic mutant)
Long considered "undruggable" (no deep pockets in static structure)
Discovery: Fragment-based crystallography combined with medicinal chemistry revealed a cryptic pocket (switch II pocket) adjacent to Cys12
AlphaFold now enables similar analyses for proteins without experimental structures
Result: Sotorasib (FDA-approved 2021), Adagrasib (FDA-approved 2022)
2. Homology-based druggability assessment If a homolog is druggable, AlphaFold structure can reveal if your target has a similar pocket.
3. Allosteric site discovery Sites distant from the active site that regulate function. Often missed in traditional screens.
What AlphaFold Doesn't Give You
1. Holo (ligand-bound) conformations AlphaFold is trained on apo structures. Induced fit and cryptic pockets are missed.
Workaround: Use experimental holo structures of homologs as templates, or run MD simulations.
2. Binding site identification AlphaFold predicts structure, not function. You still need to identify which pocket to target.
Solution: Use binding site prediction tools (FPocket, Orbion) or evolutionary conservation analysis.
3. Protein dynamics Static structure doesn't capture breathing, domain motions, or transient states.
Solution: Molecular dynamics or enhanced sampling (metadynamics, accelerated MD).
Where Orbion Fits in the SBDD Workflow
While AlphaFold provides the structure, Orbion adds the biological and functional context needed for effective drug discovery:
1. Binding Site Prediction & Ranking
What Orbion does:
Analyzes structure for potential binding sites
Scores sites by druggability (size, shape, conservation)
Identifies allosteric sites based on evolutionary conservation
Why it matters: Not all pockets are functional. Orbion filters out non-functional pockets and highlights those likely to regulate protein activity.
Example workflow:
Upload AlphaFold structure of kinase
Orbion identifies ATP pocket (expected) + allosteric site in hinge region (unexpected)
Virtual screening targets both sites
Allosteric site provides selectivity advantage
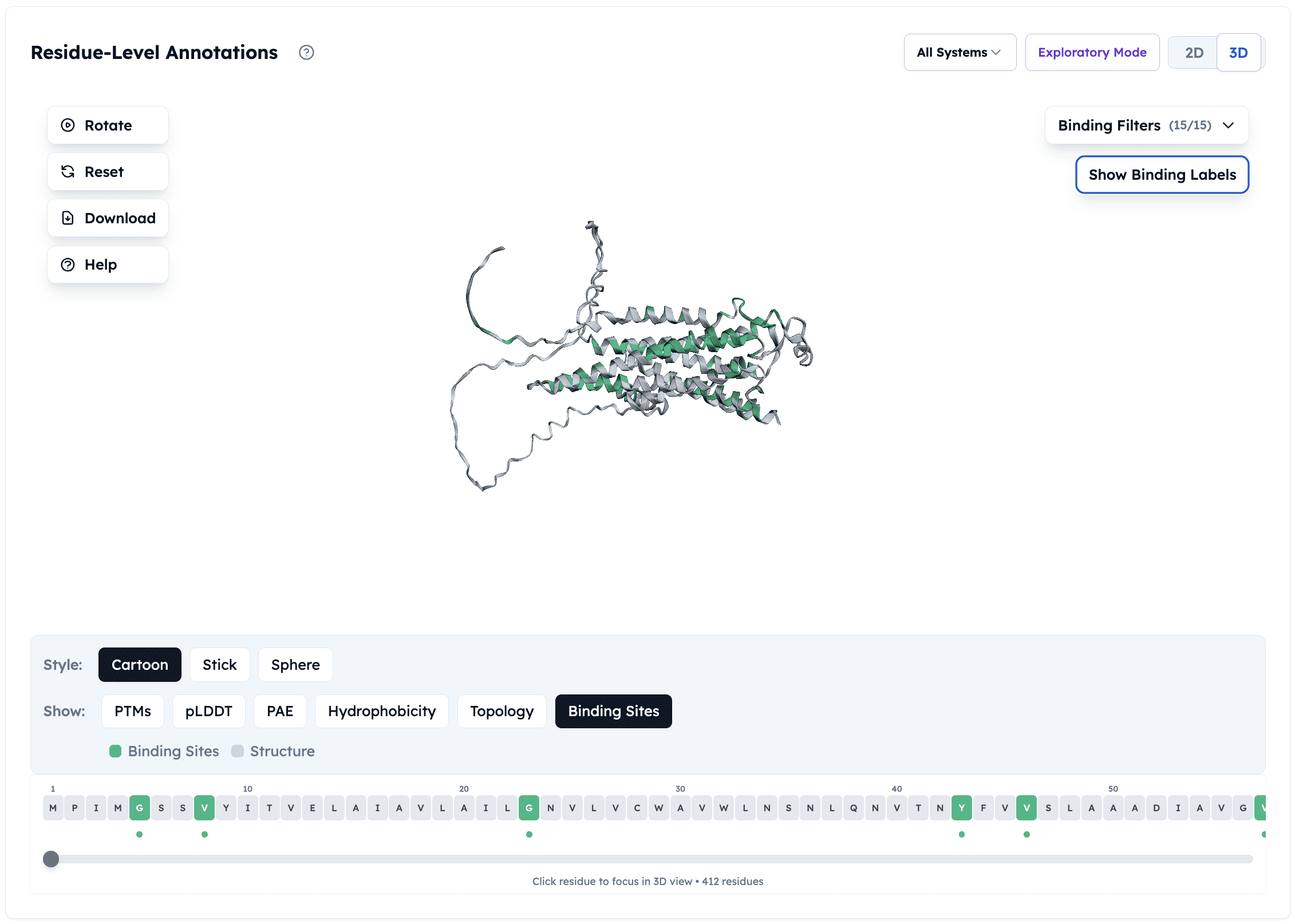
Orbion platform's binding site predictions on Adenosine Receptor A2a
2. PTM-Aware Drug Design
What Orbion does:
Predicts post-translational modifications (phosphorylation, glycosylation, ubiquitination)
Maps PTM sites onto structure
Why it matters: PTMs regulate protein activity and localization. A drug targeting the inactive state might fail if the cell modifies the protein to the active state.
Example: Kinases are activated by phosphorylation. If your AlphaFold model is unphosphorylated (inactive), but the cellular target is phosphorylated (active), your docking predictions will be wrong.
Orbion solution: Predicts phosphorylation sites, allows you to model both states.
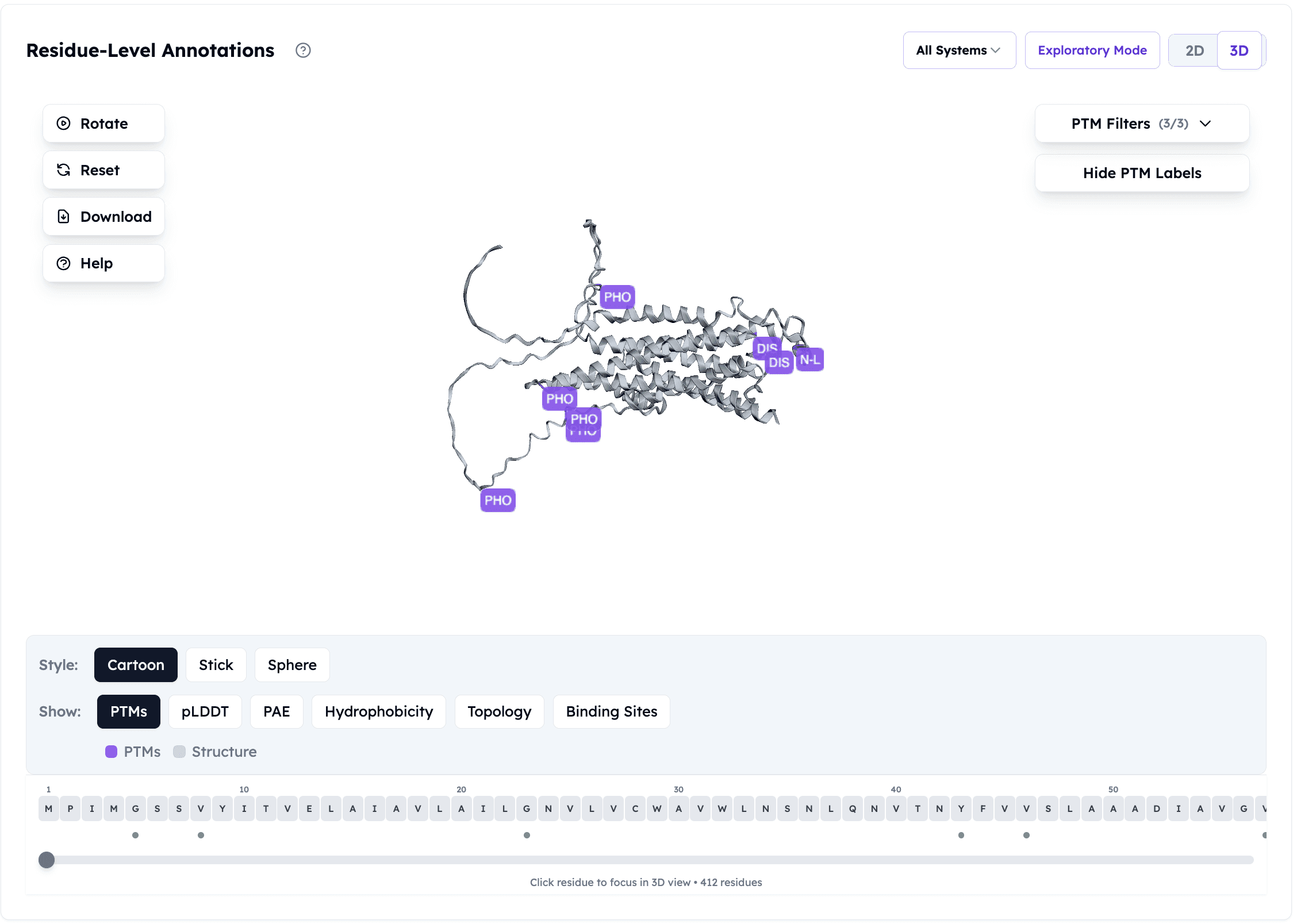
Orbion platform's post-translational modification predictions on Adenosine Receptor A2a
3. Stability & Expression Optimization
What Orbion does:
Suggests thermostabilizing mutations for construct design
Recommends expression system (E. coli, yeast, insect, mammalian)
Why it matters: To validate computational hits experimentally, you need protein. If your target is unstable or doesn't express, you can't run biochemical assays.
Example: GPCR is computationally predicted to have a druggable allosteric site, but it aggregates during purification.
Orbion solution: Suggests stabilizing mutations (from construct design module), enabling expression of stable protein for screening.
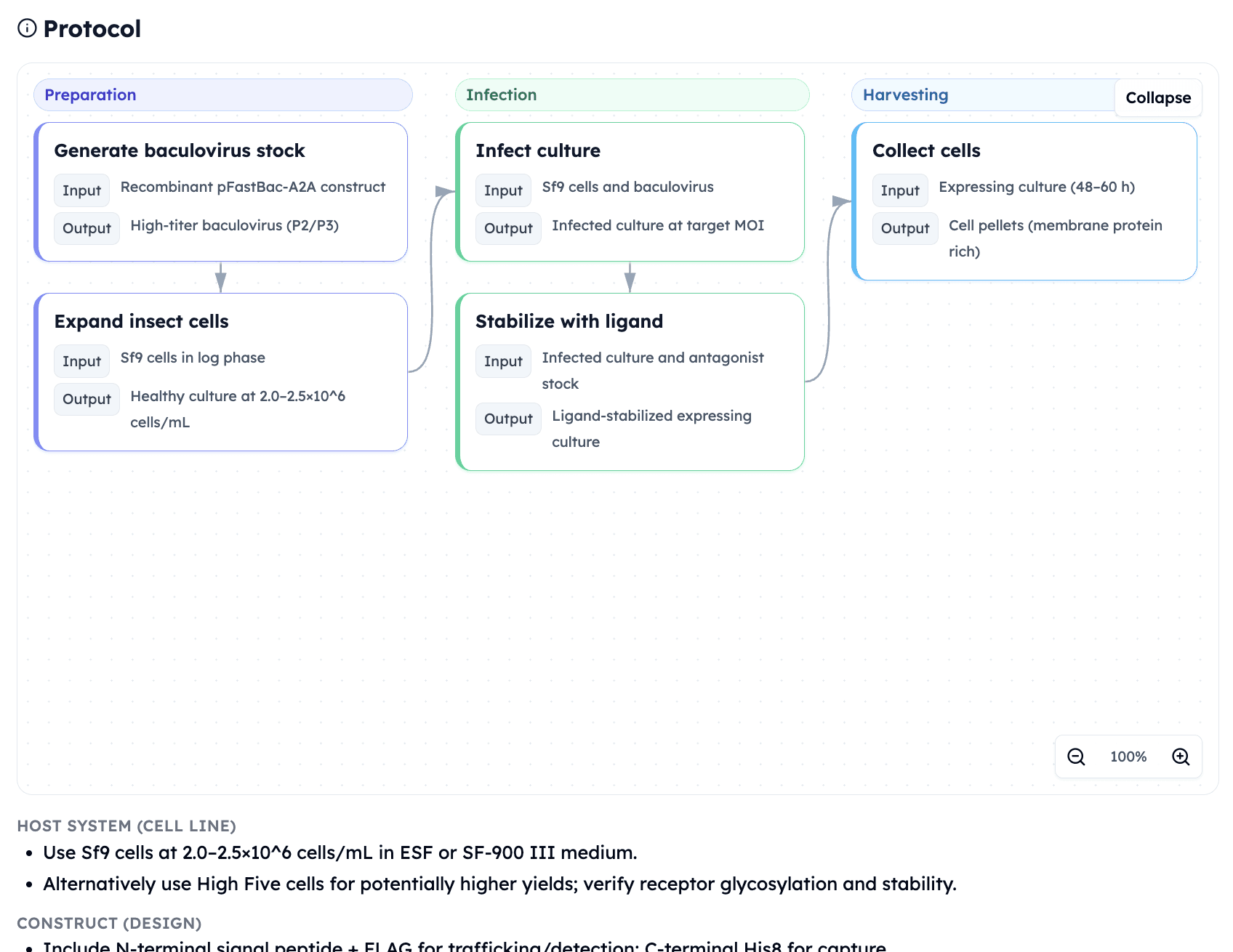
Orbion platform's expression protocol suggestion on Adenosine Receptor A2a
The Economics of SBDD
Traditional Drug Discovery Timeline
Target identification: 1-2 years
Hit discovery (HTS): 1-2 years (test 10⁶ compounds)
Lead optimization: 2-4 years
Preclinical: 1-2 years
Clinical trials: 5-10 years
Total: 10-15 years, $1-2 billion
SBDD Impact
Hit discovery: Reduced to 6-12 months (virtual screening before HTS)
Lead optimization: Faster (structure-guided modifications)
Attrition reduction: Better selectivity and ADMET early
ROI: SBDD reduces costs by 30-50% in early stages, increases probability of success by 2-3×.
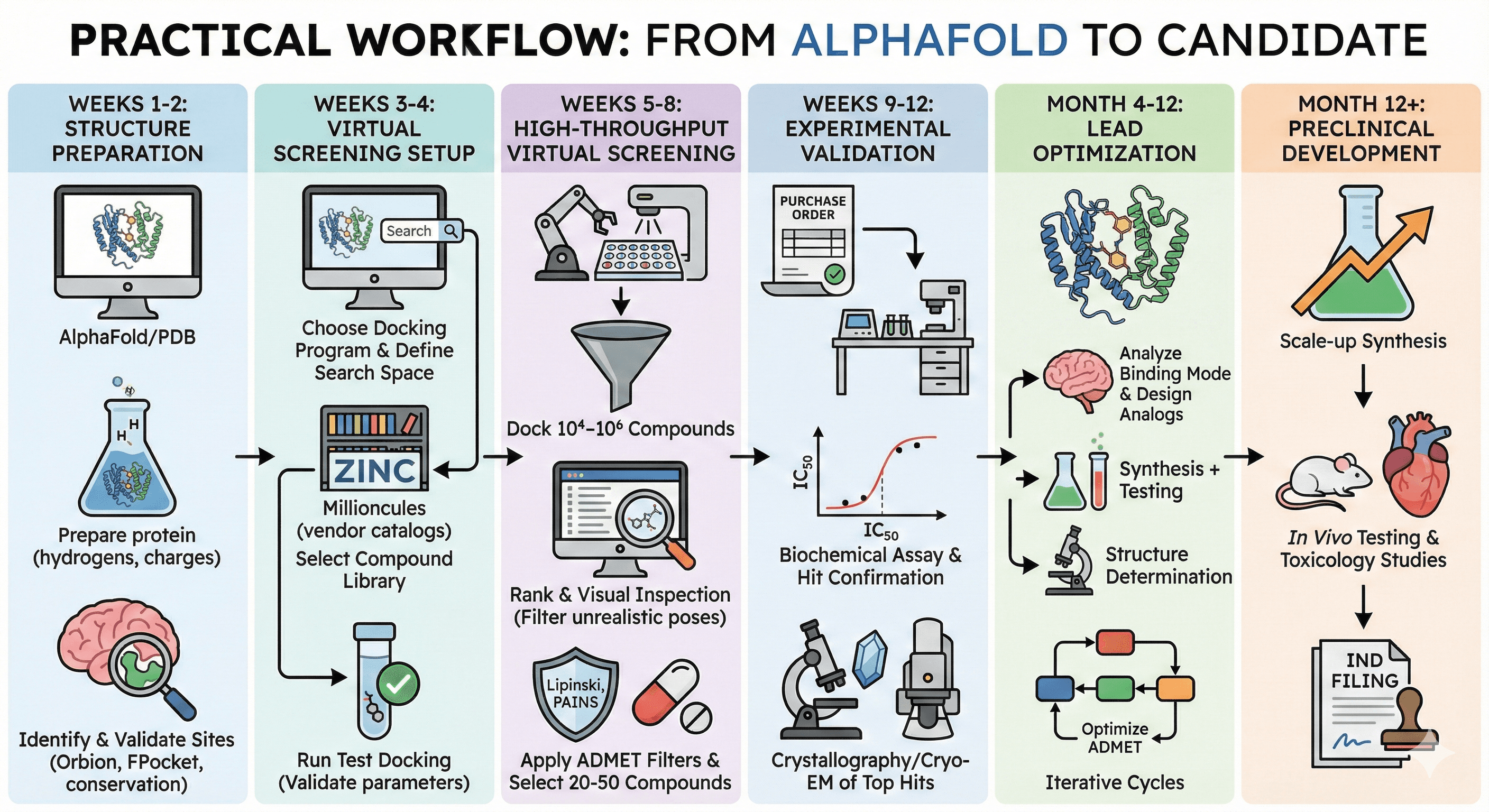
Practical Workflow: From AlphaFold to Candidate
Here's a realistic, step-by-step workflow for a structural biologist or medicinal chemist:
Week 1-2: Structure Preparation
Obtain AlphaFold structure or retrieve from PDB
Prepare protein (add hydrogens, assign charges, check for errors)
Identify binding sites (Orbion, FPocket, or manual inspection)
Validate site by checking conservation and known ligands in homologs
Week 3-4: Virtual Screening Setup
Choose docking program (AutoDock Vina for free, Glide if you have license)
Define search space (binding site box)
Select compound library (ZINC, vendor catalogs, or fragment library)
Run test docking with known ligands (if available) to validate parameters
Week 5-8: High-Throughput Virtual Screening
Dock 10⁴–10⁶ compounds
Rank by docking score
Visual inspection of top 100-500 poses (filter out unrealistic binding modes)
Apply ADMET filters (Lipinski, PAINS filters, synthesizability)
Select 20-50 compounds for experimental testing
Week 9-12: Experimental Validation
Purchase or synthesize compounds
Biochemical assay (IC₅₀ determination)
Hit confirmation (repeat, dose-response curves)
Crystallography or Cryo-EM of top hits bound to protein
Month 4-12: Lead Optimization
Solve structure of hit-protein complex
Analyze binding mode (which residues interact? Can we improve?)
Design analogs (add H-bonds, fill sub-pockets, improve selectivity)
Iterative rounds of synthesis + testing + structure determination
Optimize ADMET properties
Month 12+: Preclinical Development
Scale-up synthesis
In vivo testing (animal models)
Toxicology studies
Formulation development
IND filing
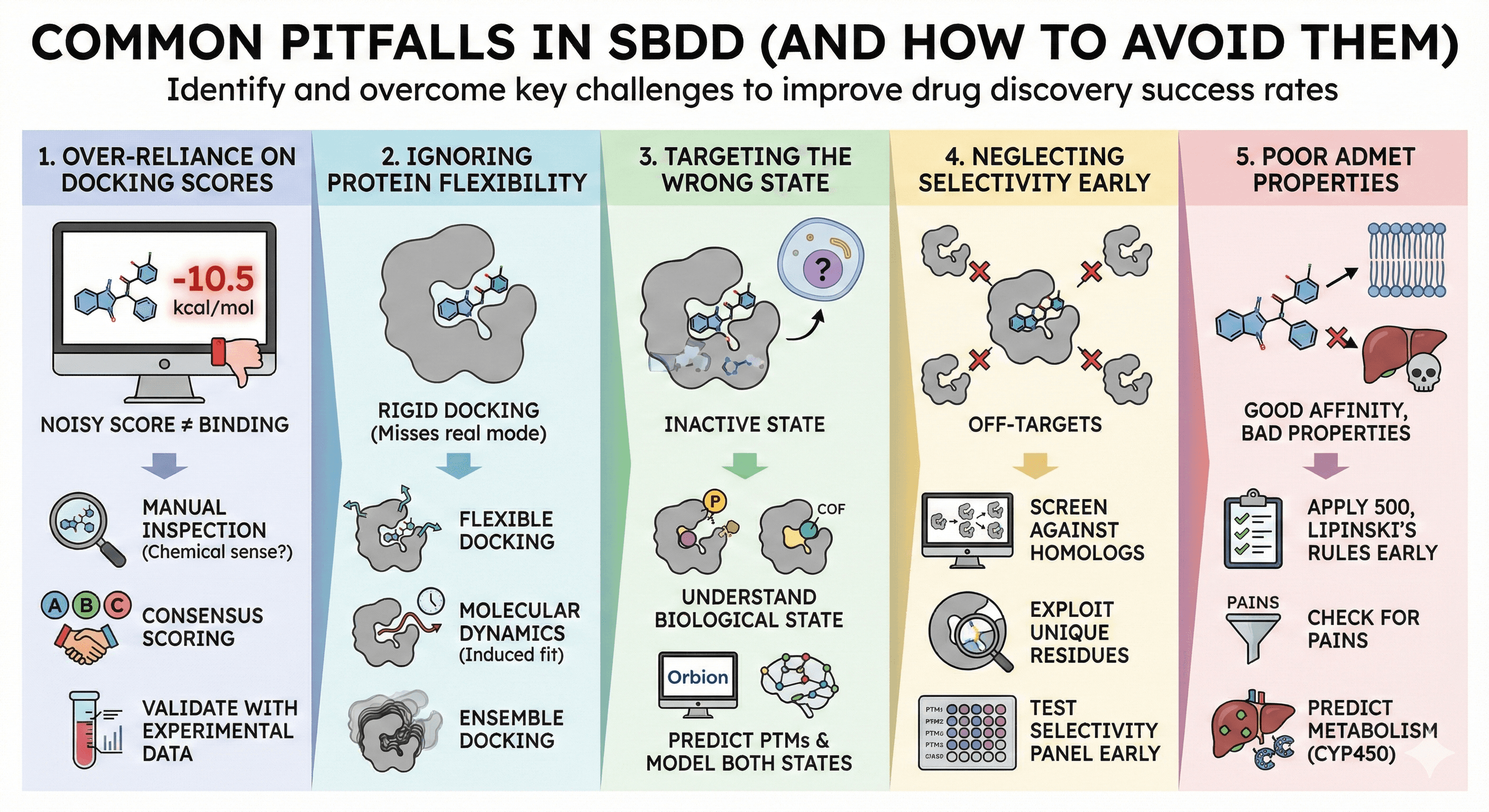
Common Pitfalls in SBDD (And How to Avoid Them)
Pitfall 1: Over-Reliance on Docking Scores
Problem: Docking scores are noisy. A score of -10 kcal/mol doesn't guarantee binding.
Solution:
Manually inspect poses (does the binding mode make chemical sense?)
Use consensus scoring (multiple programs)
Validate with experimental data (even a few known binders helps calibrate)
Pitfall 2: Ignoring Protein Flexibility
Problem: Proteins move. A rigid docking may miss the real binding mode.
Solution:
Use flexible docking (allow side chains to move)
Run molecular dynamics after docking (induced fit refinement)
Ensemble docking (dock to multiple protein conformations)
Pitfall 3: Targeting the Wrong State
Problem: You optimize a drug for the inactive state, but the cell has the active state.
Solution:
Understand the biological state (is the protein phosphorylated? Bound to a cofactor?)
Use Orbion to predict PTMs and model both states
Check if homologs have structures in active/inactive states
Pitfall 4: Neglecting Selectivity Early
Problem: You find a potent inhibitor, but it hits 50 other kinases.
Solution:
Screen against homologs computationally (docking to off-targets)
Exploit unique residues in your target's binding site
Test selectivity panel early (before investing in optimization)
Pitfall 5: Poor ADMET Properties
Problem: Great affinity, but it doesn't cross membranes or is metabolized instantly.
Solution:
Apply Lipinski's Rule of Five early (MW < 500, LogP < 5, H-bond donors < 5, acceptors < 10)
Check for PAINS (pan-assay interference compounds)
Predict metabolism (CYP450 interactions)
The Future of SBDD: AI-Native Drug Design
The next generation of SBDD doesn't just use AI for structure prediction—it uses AI for molecule generation.
Generative Models for Drug Design
Traditional: Dock existing compounds → Pick best
AI-Native: Generate novel compounds optimized for the binding site
Tools emerging:
Insilico Medicine (Chemistry42): Generates molecules de novo for a given target
Relay Therapeutics: Combines MD simulations + AI to find allosteric sites + design binders
Exscientia: AI-designed drugs already in clinical trials (faster timelines)
How it works:
Train generative model (VAE, GAN, or Transformer) on millions of drug-like molecules
Condition generation on binding site structure
Model generates molecules predicted to bind
Filter for synthesizability and ADMET
Synthesize top candidates
Impact: First AI-designed drugs are entering clinics (e.g., DSP-1181 for OCD, designed by Exscientia in 12 months vs typical 4-5 years).
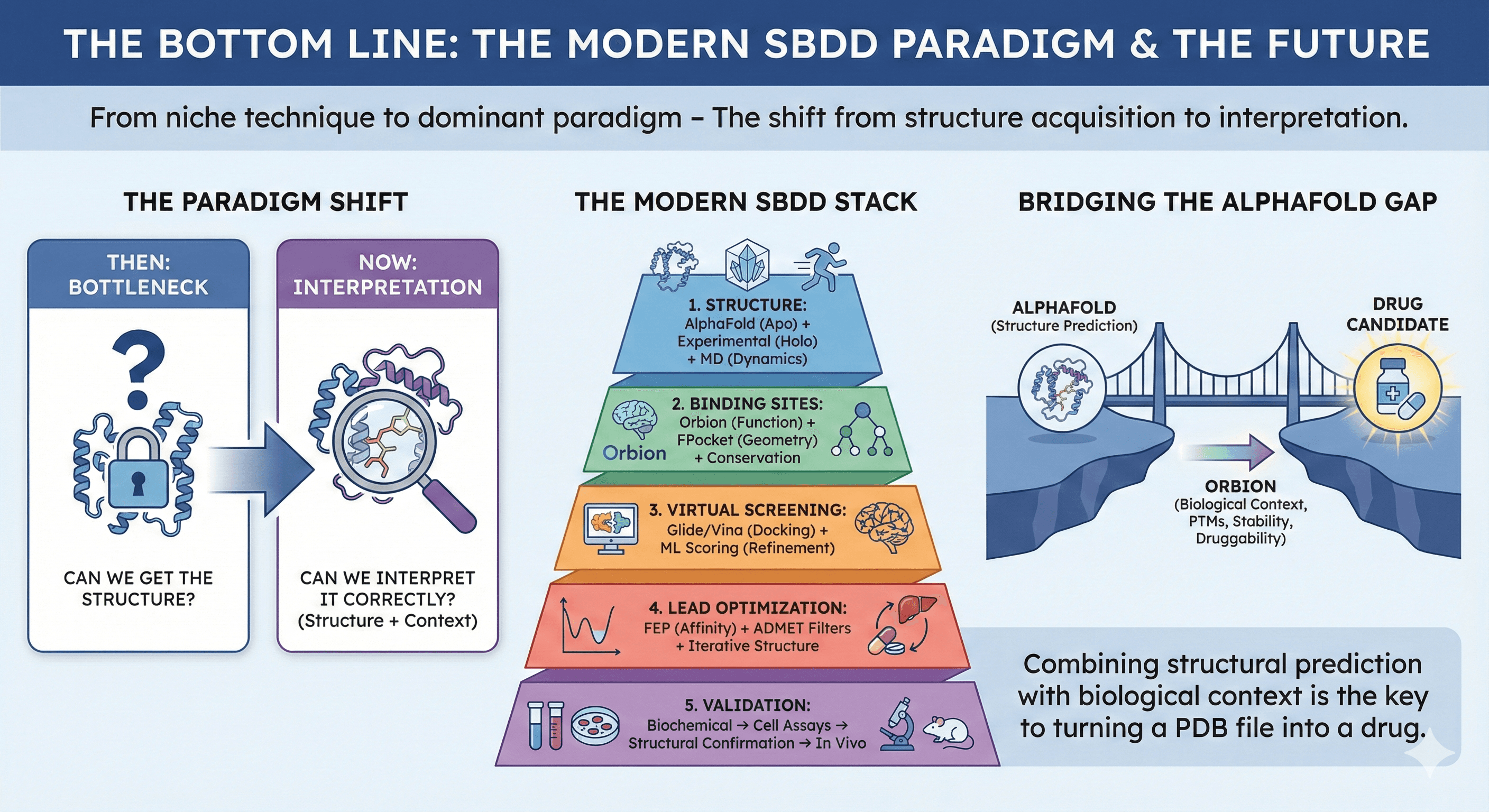
The Bottom Line
Structure-based drug design has matured from a niche technique to the dominant paradigm in drug discovery. With AlphaFold democratizing access to structures and AI tools accelerating virtual screening and lead optimization, the bottleneck is shifting from "Can we get the structure?" to "Can we interpret it correctly?"
The modern SBDD stack:
Structure: AlphaFold (apo state) + experimental structures (holo state) + MD simulations (dynamics)
Binding sites: Orbion (functional prediction) + FPocket (geometry) + evolutionary conservation
Virtual screening: Glide/Vina (docking) + Machine learning scoring (refinement)
Lead optimization: FEP (affinity prediction) + ADMET filters + iterative structure determination
Validation: Biochemical assays → Cell assays → Structural confirmation → In vivo
The gap that AlphaFold left—PTMs, binding site identification, stability, druggability assessment—is where platforms like Orbion add critical value. Combining structural prediction with biological context is the key to turning a PDB file into a drug.

