Blog
The Art of "Domesticating" Proteins: A Guide to GPCR Construct Design
Dec 10, 2025
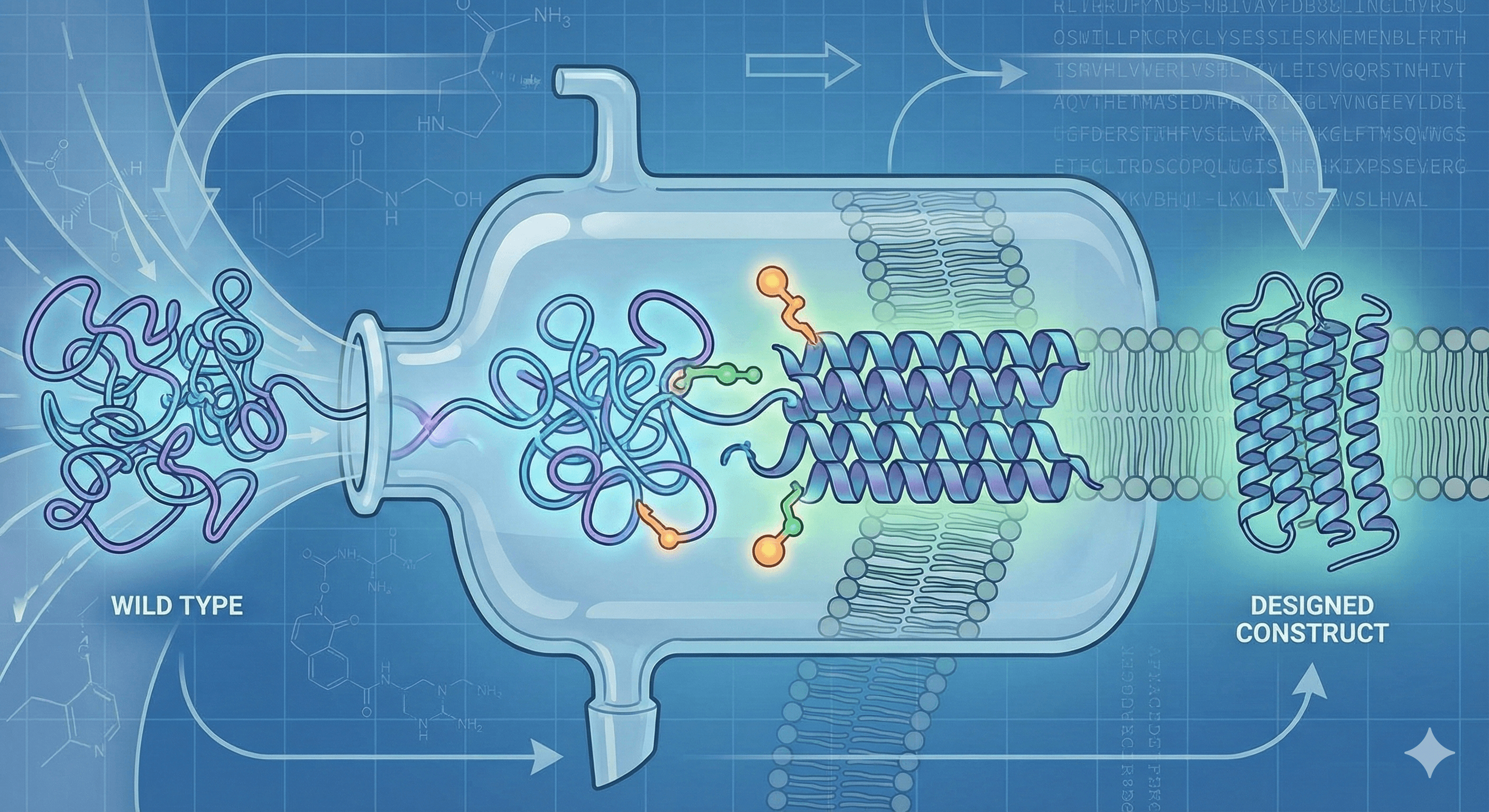
Any structural biologist knows the sinking feeling of "Expression Hell." You clone your gene, put it into HEK293 or Sf9 cells, and… nothing. Or worse, you get a goopy, aggregated mess at the bottom of a test tube.
Why? Because wild-type membrane proteins are conformational shapeshifters. They evolved for the comfortable embrace of a lipid bilayer, not the harsh reality of a detergent micelle. To study them effectively, we cannot just use the protein as nature made it. We have to "domesticate" it through Construct Design.
This process is the difference between a failed project and a high-resolution structure. Here is the three-step architecture of a stabilized GPCR construct.
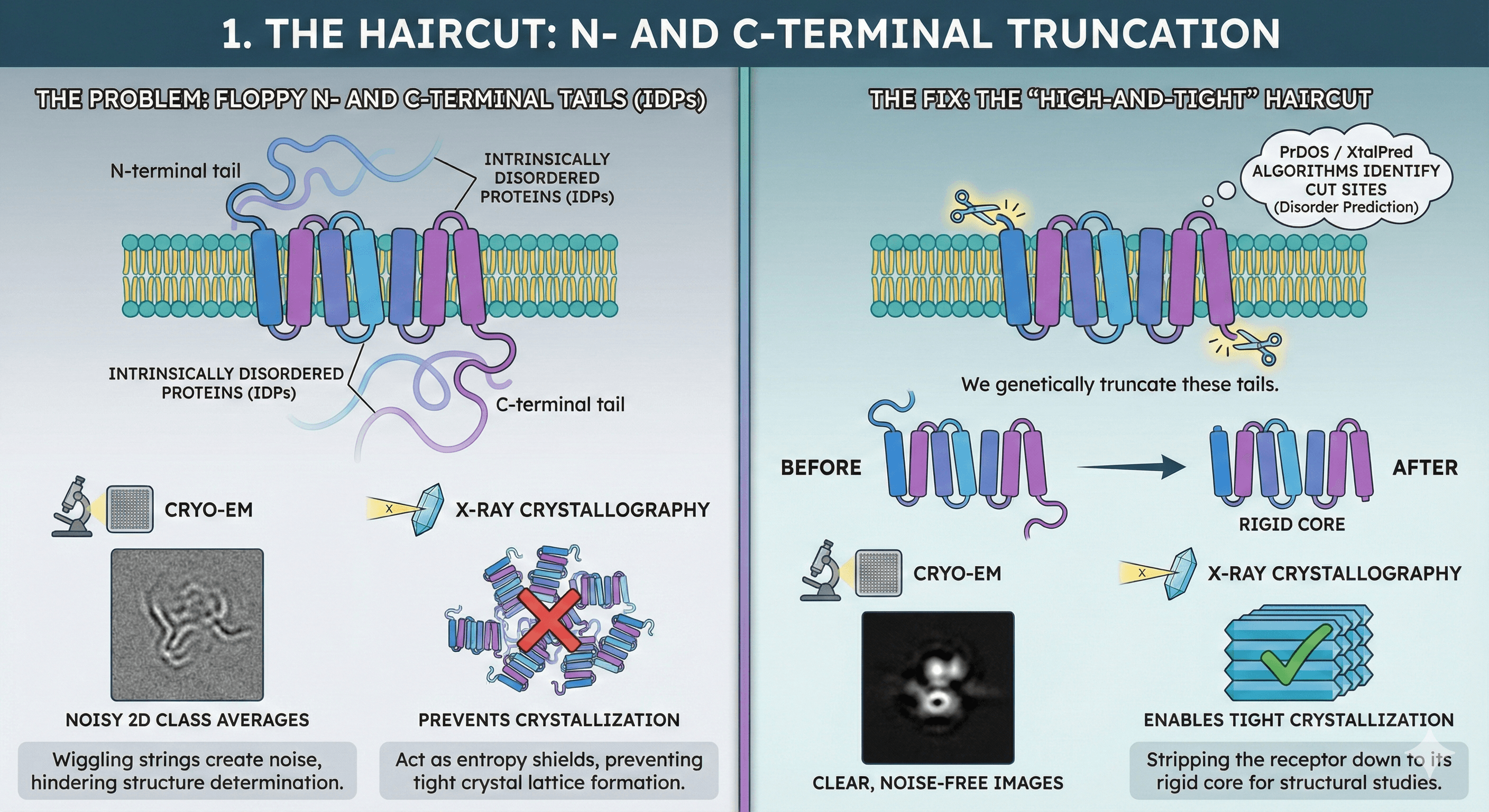
1. The Haircut: N- and C-Terminal Truncation
The Problem: Most GPCRs have long N-terminal and C-terminal tails. While biologically crucial for signaling (e.g., for β-arrestin recruitment or internalization), biophysically, these regions are usually Intrinsically Disordered Proteins (IDPs).
In Cryo-EM, these "wiggling strings" create noise in 2D class averages. In X-ray crystallography, they act like entropy shields, preventing the tight crystal lattice formation required for diffraction.
The Fix: We give the protein a "high-and-tight" haircut.
The Tool: We use disorder prediction algorithms like PrDOS or XtalPred to identify exactly where the structured 7TM bundle ends and the disorder begins.
The Action: We genetically truncate these tails, stripping the receptor down to its rigid core.
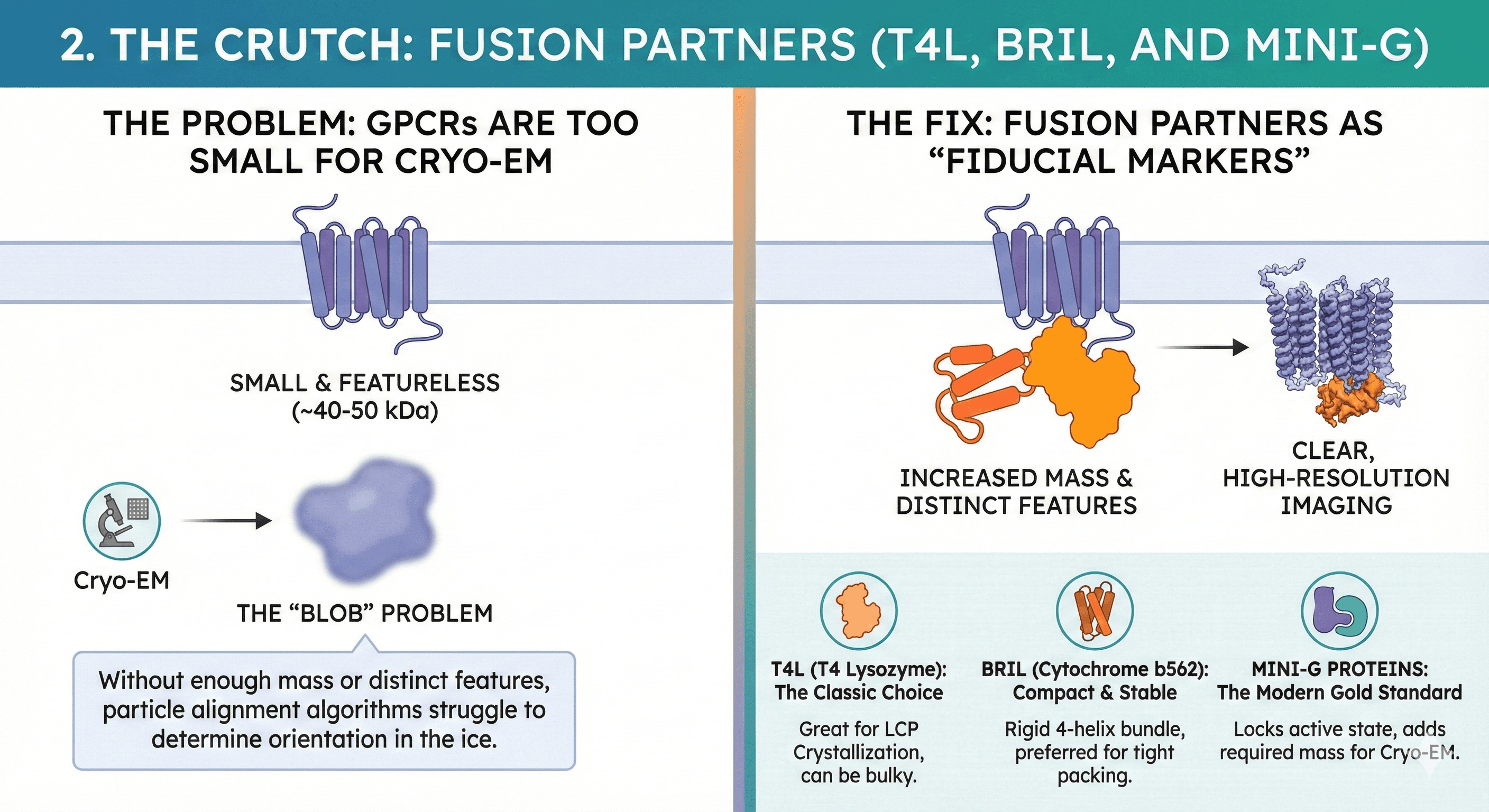
2. The Crutch: Fusion Partners (T4L, BRIL, and Mini-G)
The Problem: GPCRs are relatively small (~40-50 kDa). In the world of Cryo-EM, small is bad. Without enough mass or distinct features, particle alignment algorithms struggle to determine the orientation of the protein in the ice (the "blob" problem).
The Fix: We insert a rigid, soluble fusion protein to serve as a "fiducial marker"—a distinct shape for the microscope to lock onto. This is typically fused into the 3rd Intracellular Loop (ICL3) or attached to the N-terminus.
T4 Lysozyme (T4L): The classic choice that enabled the first GPCR crystal structures. It is excellent for Lipid Cubic Phase (LCP) crystallization but can sometimes be too bulky for certain conformations.
BRIL (Cytochrome b562): A highly stable, 4-helix bundle. It is more rigid and compact than T4L, often preferred when the fusion needs to sit tightly against the receptor without disrupting the fold.
Mini-G Proteins: The modern gold standard for Cryo-EM. By fusing a truncated, engineered G-protein to the receptor, we can lock the GPCR in its active state while simultaneously adding the mass needed for high-resolution imaging.
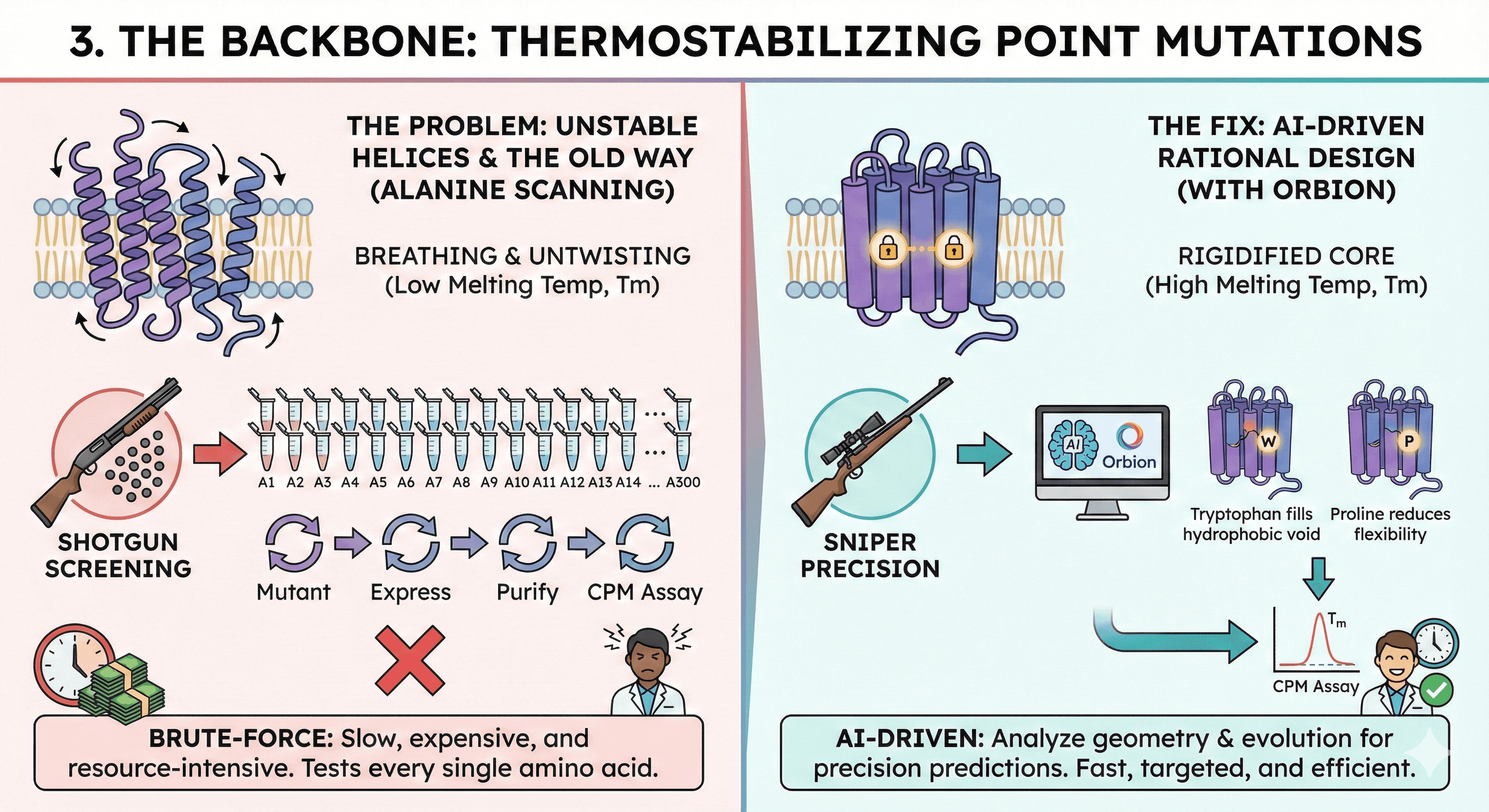
3. The Backbone: Thermostabilizing Point Mutations
The Problem: Even with a haircut and a fusion partner, the transmembrane helices can "breathe" or untwist when removed from the membrane. To get a structure, we need to raise the Melting Temperature (Tm), which is typically measured via a CPM (Cell Membrane Protein) Thermal Shift Assay.
The Fix: We introduce single amino acid changes to rigidify the core.
The Old Way (Alanine Scanning): Historically, this was a brute-force game. We would mutate every single amino acid to Alanine, one by one.
Mutant 1 → Express → Purify → CPM Assay.
Mutant 2 → Express → Purify → CPM Assay. It is effective, but it is also slow, expensive, and resource-intensive.
The New Way (Rational Design with Orbion): Today, we use AI-driven rational design. Platforms like Orbion analyze the geometry and evolutionary history of the protein to make precision predictions. Instead of testing 300 random mutants, Orbion might suggest: "Change Residue 124 to Tryptophan to fill this hydrophobic void," or "Mutate Residue 340 to Proline to reduce helix flexibility." This moves the workflow from "shotgun" screening to "sniper" precision.
Summary: The Construct Design Toolkit
For the practicing structural biologist, here is how the toolkit stacks up:
Challenge | Technical Solution | Key Tool/Reagent |
|---|---|---|
Disordered/Floppy Tails | Region Truncation | PrDOS / XtalPred |
Small Particle Size | Fusion Proteins (Fiducial Markers) | BRIL / T4L / Rubredoxin |
Conformational Instability | Thermostabilizing Mutations | Orbion (Rational Design) |
Active State Capture | Complex Stabilization | Mini-G Proteins / Nanobodies (Nb80) |
Stability Measurement | Thermal Unfolding | CPM Assay / nanoDSF |
The Takeaway
Nature builds proteins to move and signal; we build constructs to sit still and diffract.
The difference between a "failed target" and a "druggable breakthrough" is often found in the engineering. By combining the classic strategies of truncations and fusions with modern AI-driven stabilization tools like Orbion, we are finally turning the dark art of construct design into a predictable, reproducible science.