Blog
Construct Boundary Design: The 4 Problems Killing Your Protein Expression
Jan 9, 2026
You clone the full-length gene. Express it. Inclusion bodies. You try with tags. Still insoluble. You try insect cells. It expresses, but aggregates during purification. You truncate the C-terminus—guess where. The protein is now soluble but completely inactive.
Six months gone, and you still don't have a working construct.
The problem isn't your expression system or purification protocol. It's your construct boundaries. Defining where a protein starts and ends—what to include, what to truncate—is the single most important decision in structural biology.
Key Takeaways
80-90% of construct failures are due to wrong boundaries
Main problems: Disordered termini (40%), flexible internal loops (20%), wrong domain boundaries (30%), bad tag placement (10%)
Average constructs tested: 5-15 per successful structure (traditional trial-and-error)
Cost of poor boundaries: $50-100K wasted per failed construct (6-12 months)
Modern solution: AI-driven boundary prediction reduces constructs tested from 10+ to 1-3
Success rate improvement: 60-80% first-construct success (vs 15-25% traditional)

What Are Construct Boundaries?
Construct boundaries define which residues you express and purify:
N-terminal boundary: Where does your protein start?
C-terminal boundary: Where does it end?
Internal boundaries: Which loops or domains do you include, truncate, or replace?
Example:
Full-length: Residues 1-450
Your construct: Residues 28-412 (N-terminal truncation of 27 residues, C-terminal truncation of 38 residues)

Why Boundaries Matter: The Goldilocks Principle
Include too much (disordered regions):
Flexible termini prevent crystallization
Disordered loops cause aggregation
PTMs create heterogeneity (blocks cryo-EM)
Result: Expresses but won't crystallize/freeze
Include too little (remove essential domains):
Protein loses stability (unfolds, aggregates)
Loses function (active site disrupted)
Result: Doesn't express, or is insoluble/inactive
Just right (structured core, minimal disorder):
Protein is stable, monodisperse
Crystallizes or forms good cryo-EM grids
Retains function
Result: Structure determination succeeds
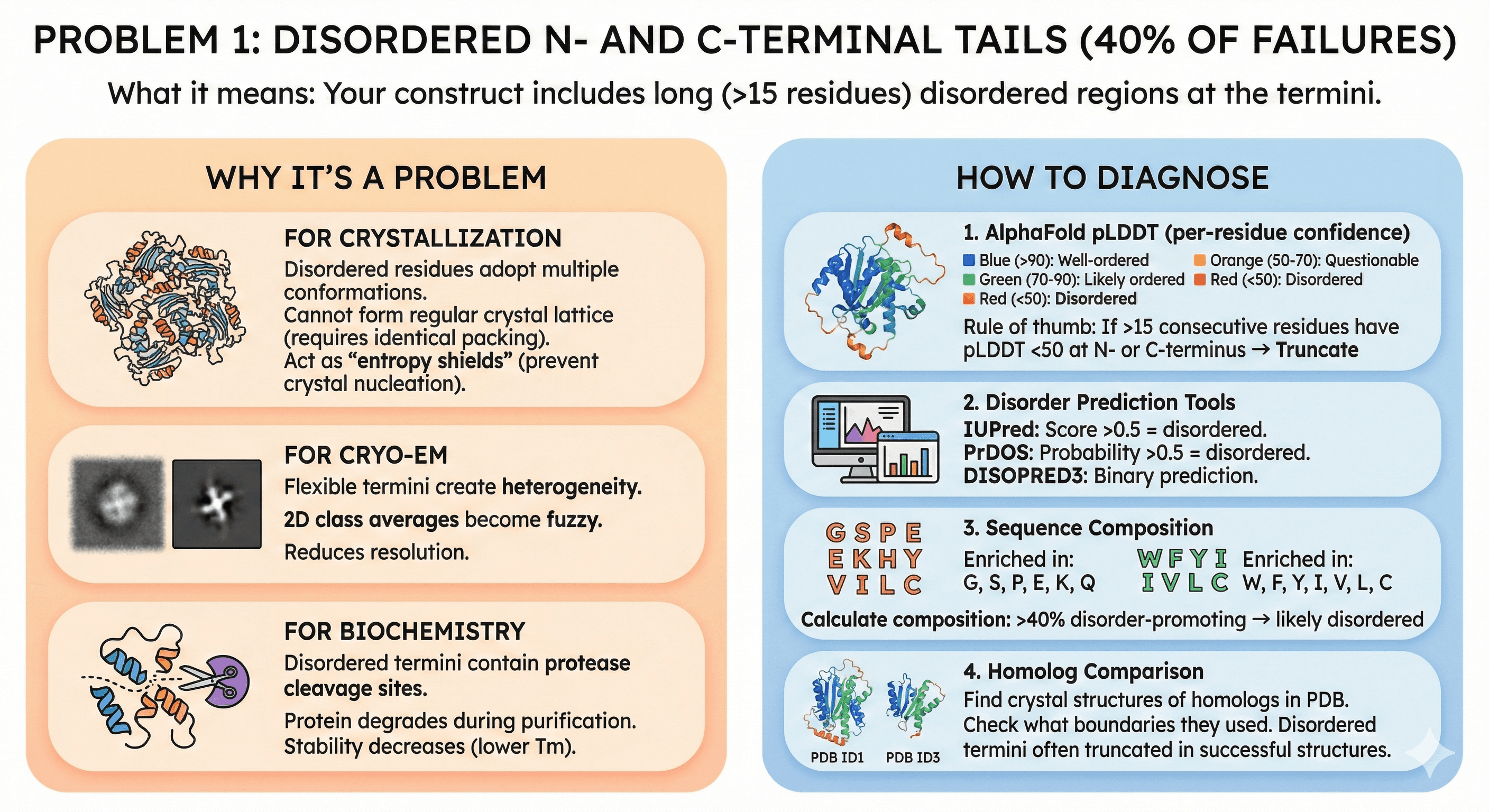
Problem 1: Disordered N- and C-Terminal Tails (40% of Failures)
What it means: Your construct includes long (>15 residues) disordered regions at the termini.
Why It's a Problem
For crystallization:
Disordered residues adopt multiple conformations
Cannot form regular crystal lattice (requires identical packing)
Act as "entropy shields" (prevent crystal nucleation)
For cryo-EM:
Flexible termini create heterogeneity
2D class averages become fuzzy
Reduces resolution
For biochemistry:
Disordered termini contain protease cleavage sites
Protein degrades during purification
Stability decreases (lower Tm)
How to Diagnose
1. AlphaFold pLDDT (per-residue confidence):
Blue (pLDDT >90): Well-ordered
Green (pLDDT 70-90): Likely ordered
Orange (pLDDT 50-70): Questionable
Red (pLDDT <50): Disordered
Rule of thumb: If >15 consecutive residues have pLDDT <50 at N- or C-terminus → Truncate
2. Disorder prediction tools:
IUPred: Score >0.5 = disordered
PrDOS: Probability >0.5 = disordered
DISOPRED3: Binary prediction
3. Sequence composition:
Disordered regions enriched in: Gly, Ser, Pro, Glu, Lys, Gln
Structured regions enriched in: Trp, Phe, Tyr, Ile, Val, Leu, Cys
Calculate composition: >40% disorder-promoting → likely disordered
4. Homolog comparison:
Find crystal structures of homologs in PDB
Check what boundaries they used
Disordered termini often truncated in successful structures
Example: Bacterial Enzyme
Full-length (1-380):
AlphaFold pLDDT:
Residues 1-22: pLDDT 30-50 (disordered)
Residues 23-355: pLDDT 85-95 (structured)
Residues 356-380: pLDDT 40-60 (disordered)
Test constructs:
Construct 1 (1-380, full-length): Expresses, won't crystallize
Construct 2 (23-355, truncate both): Expresses, crystallizes, 2.3 Å
Construct 3 (30-350, aggressive): Expresses, lower yield/stability
Winner: Construct 2 (remove disorder, keep structured core)
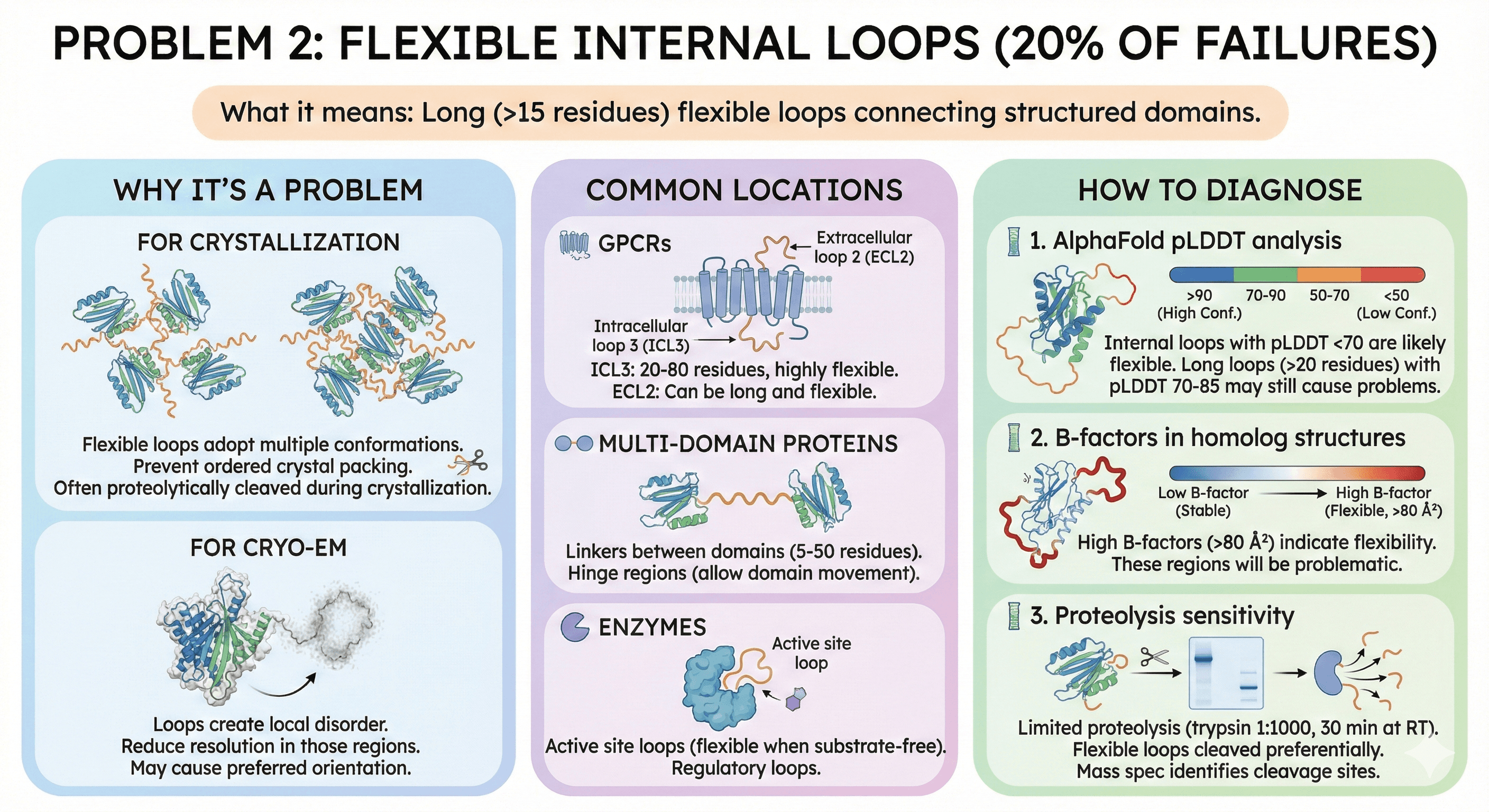
Problem 2: Flexible Internal Loops (20% of Failures)
What it means: Long (>15 residues) flexible loops connecting structured domains.
Why It's a Problem
For crystallization:
Flexible loops adopt multiple conformations
Prevent ordered crystal packing
Often proteolytically cleaved during crystallization
For cryo-EM:
Loops create local disorder
Reduce resolution in those regions
May cause preferred orientation
Common Locations
GPCRs:
Intracellular loop 3 (ICL3): 20-80 residues, highly flexible
Extracellular loop 2 (ECL2): Can be long and flexible
Multi-domain proteins:
Linkers between domains (5-50 residues)
Hinge regions (allow domain movement)
Enzymes:
Active site loops (flexible when substrate-free)
Regulatory loops
How to Diagnose
1. AlphaFold pLDDT analysis:
Internal loops with pLDDT <70 are likely flexible
Long loops (>20 residues) with pLDDT 70-85 may still cause problems
2. B-factors in homolog structures:
High B-factors (>80 Ų) indicate flexibility
These regions will be problematic
3. Proteolysis sensitivity:
Limited proteolysis (trypsin 1:1000, 30 min at RT)
Flexible loops cleaved preferentially
Mass spec identifies cleavage sites
The GPCR ICL3 Problem
Famous example: β2-adrenergic receptor
ICL3 (residues ~230-270): 40 residues, highly flexible
Wild-type: Cannot crystallize
Solution: Replace ICL3 with T4 lysozyme
Construct: Residues 1-229 + T4L + 271-350
Result: First GPCR crystal structure (2007, Nobel Prize 2012)
Why it worked:
T4L is rigid, well-behaved
Provides crystal contacts
Acts as "fiducial marker" for alignment
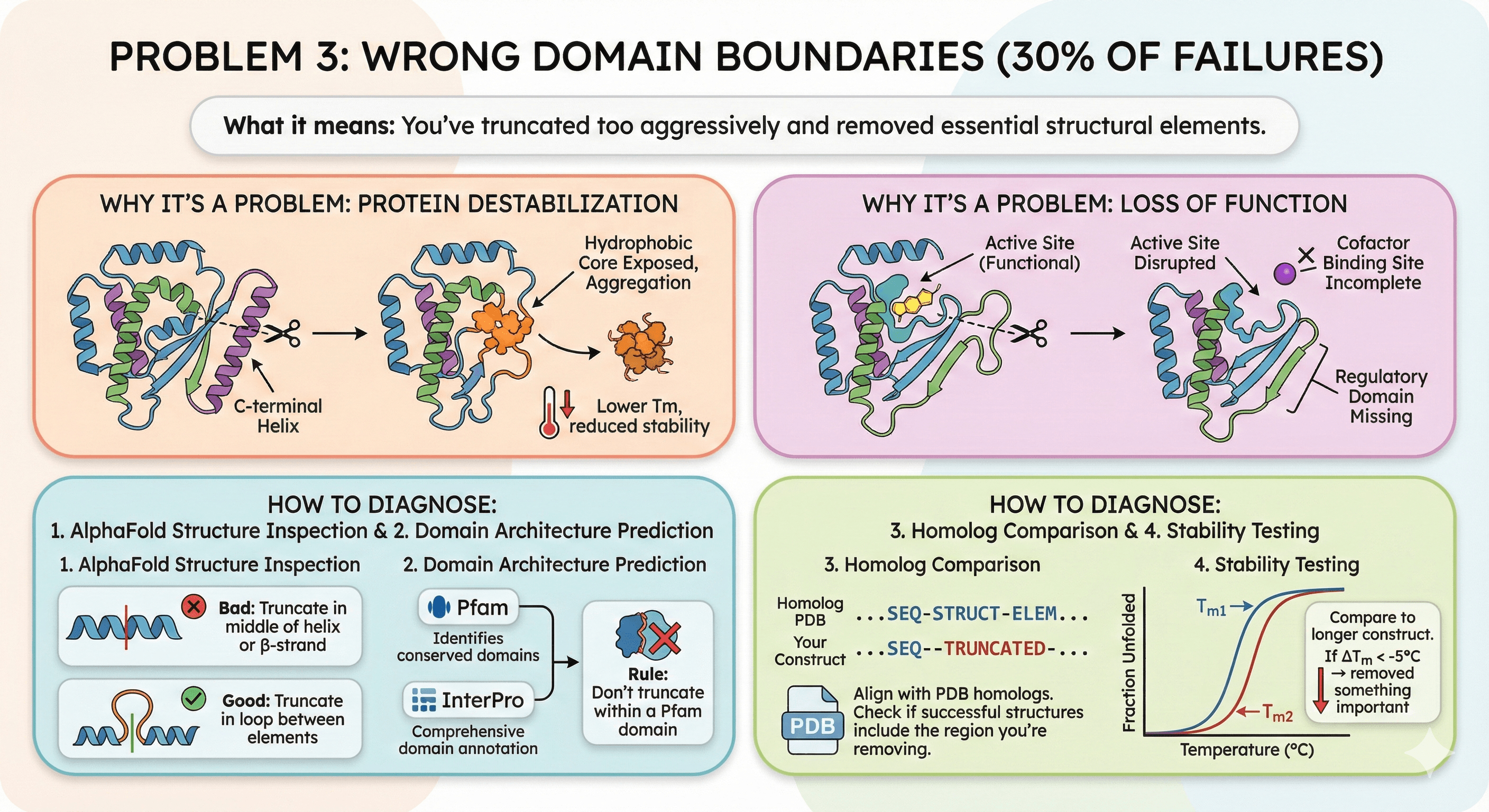
Problem 3: Wrong Domain Boundaries (30% of Failures)
What it means: You've truncated too aggressively and removed essential structural elements.
Why It's a Problem
Protein destabilization:
Removing C-terminal helix exposes hydrophobic core
Protein unfolds, aggregates
Lower Tm, reduced stability
Loss of function:
Active site disrupted
Cofactor binding site incomplete
Regulatory domain missing
How to Diagnose
1. AlphaFold structure inspection:
Check if truncation cuts through secondary structure
Bad: Truncate in middle of helix or β-strand
Good: Truncate in loop between elements
2. Domain architecture prediction:
Pfam: Identifies conserved domains
InterPro: Comprehensive domain annotation
Rule: Don't truncate within a Pfam domain
3. Homolog comparison:
Align with PDB homologs
Check if successful structures include the region you're removing
4. Stability testing:
Express truncated construct, measure Tm
Compare to longer construct
If ΔTm < -5°C → removed something important
Case Study: Removing Essential Helix
Target: Novel bacterial enzyme
Attempt 1:
Construct: Residues 1-320 (removed C-terminal 30 residues)
Expression: Inclusion bodies (completely insoluble)
Analysis:
AlphaFold structure: Residues 310-330 form amphipathic helix
Helix packs against core (stabilizes hydrophobic pocket)
Removing it exposes core → aggregation
Attempt 2:
Construct: Residues 1-340 (keep helix, remove last 10 disordered)
Expression: Soluble, stable
Crystallization: Success, 2.1 Å structure
Lesson: Even one helix can be critical. Respect secondary structure boundaries.
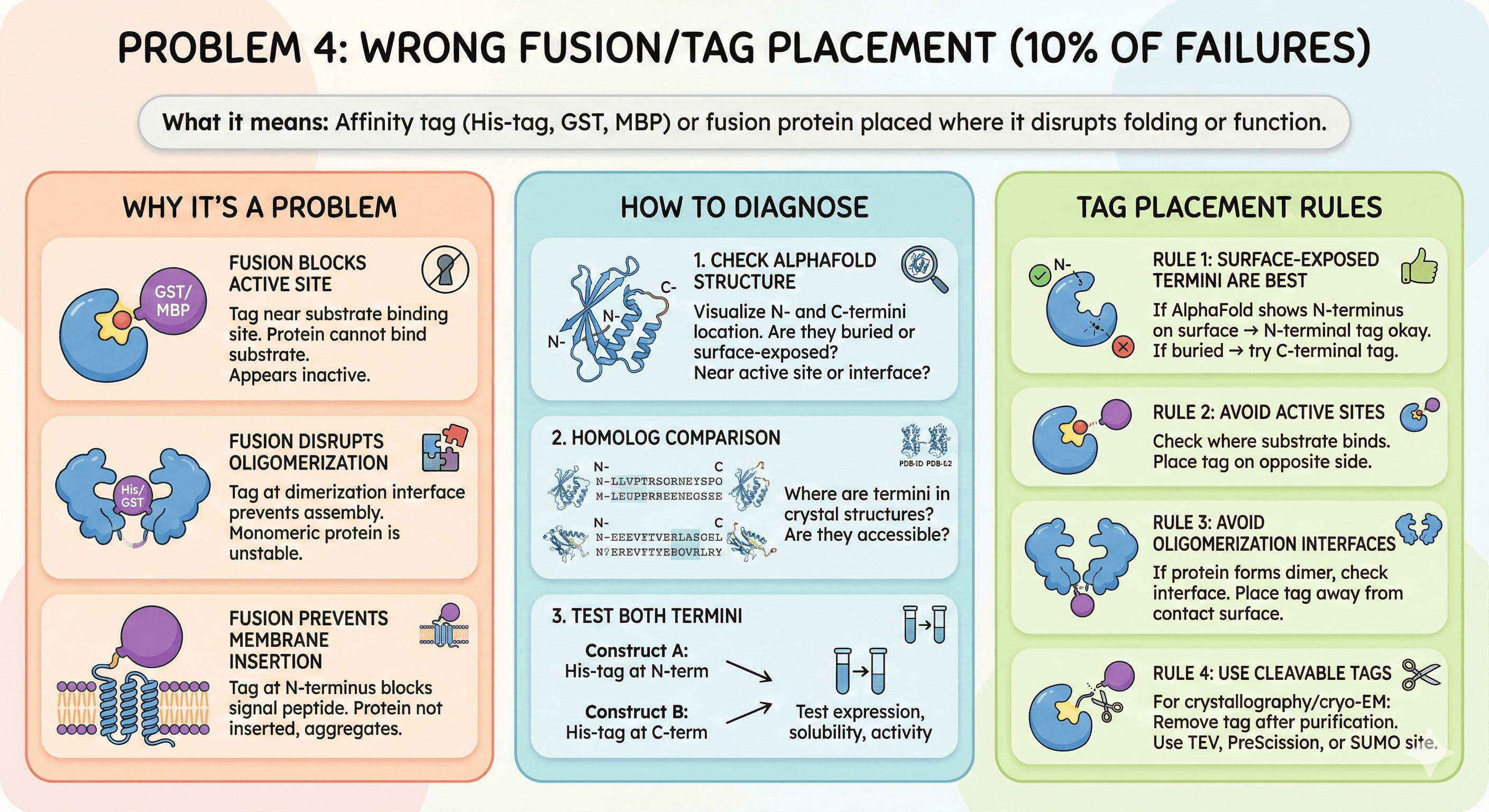
Problem 4: Wrong Fusion/Tag Placement (10% of Failures)
What it means: Affinity tag (His-tag, GST, MBP) or fusion protein placed where it disrupts folding or function.
Why It's a Problem
Fusion blocks active site:
Tag near substrate binding site
Protein cannot bind substrate
Appears inactive
Fusion disrupts oligomerization:
Protein normally forms dimer
Tag at dimerization interface prevents assembly
Monomeric protein is unstable
Fusion prevents membrane insertion:
For membrane proteins, N-terminal signal peptide required
Tag at N-terminus blocks signal peptide
Protein not inserted, aggregates
How to Diagnose
1. Check AlphaFold structure:
Visualize where N- and C-termini are located
Are they buried or surface-exposed?
Are they near active site or oligomerization interface?
2. Homolog comparison:
Where are termini in crystal structures?
Are they accessible?
3. Test both termini:
Construct A: His-tag at N-terminus
Construct B: His-tag at C-terminus
Test expression, solubility, activity
Tag Placement Rules
Rule 1: Surface-exposed termini are best
If AlphaFold shows N-terminus on surface → N-terminal tag okay
If buried → try C-terminal tag
Rule 2: Avoid active sites
Check where substrate binds
Place tag on opposite side
Rule 3: Avoid oligomerization interfaces
If protein forms dimer, check interface
Place tag away from contact surface
Rule 4: Use cleavable tags
For crystallography/cryo-EM: Remove tag after purification
Use TEV protease site, PreScission site, or SUMO
Multi-Domain Proteins: Special Considerations
Example: Full-length (1-520)
Domain 1 (catalytic): Residues 45-280
Linker (flexible): Residues 281-310
Domain 2 (regulatory): Residues 311-490
C-terminal tail (disordered): Residues 491-520
Good construct options:
Construct A: 45-280 (catalytic domain only)
Construct B: 45-490 (both domains, no disordered tail)
Construct C: 311-490 (regulatory domain only)
Bad construct options:
Construct D: 45-250 (truncates within catalytic domain) → Unstable
Construct E: 100-490 (removes part of catalytic domain) → Inactive
Key principle: Keep entire domains, remove linkers between them.
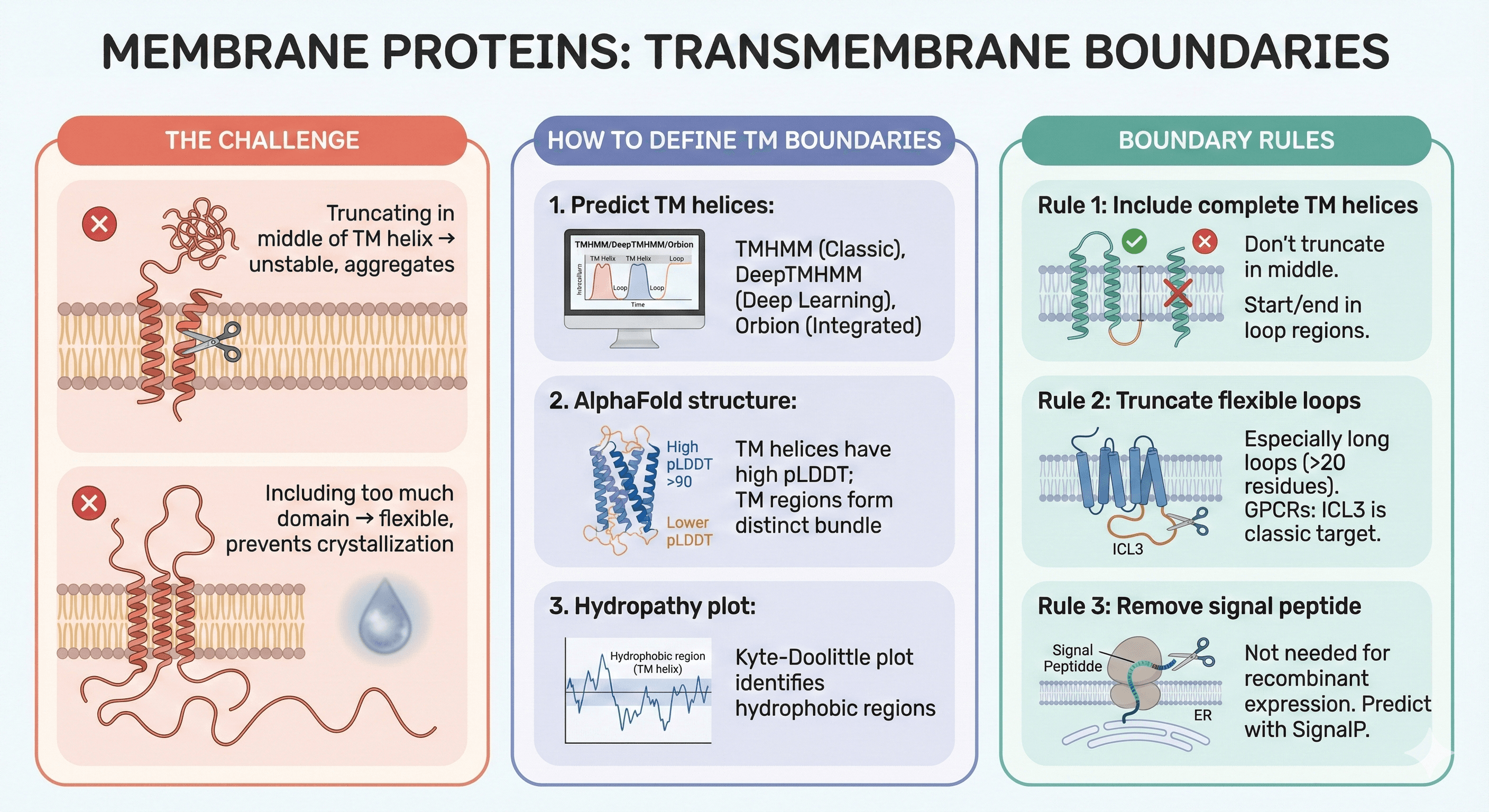
Membrane Proteins: Transmembrane Boundaries
The Challenge
Need to include entire transmembrane (TM) helices
Truncating in middle of TM helix → unstable, aggregates
Including too much intracellular/extracellular domain → flexible, prevents crystallization
How to Define TM Boundaries
1. Predict TM helices:
TMHMM: Classic tool, reliable
DeepTMHMM: Deep learning-based, more accurate
Orbion: Integrates TM prediction with structure
2. AlphaFold structure:
TM helices have high pLDDT (usually >90)
TM regions form distinct bundle
3. Hydropathy plot:
Kyte-Doolittle plot identifies hydrophobic regions (TM helices)
Boundary Rules for Membrane Proteins
Rule 1: Include complete TM helices
Don't truncate in middle of helix
Start/end in loop regions
Rule 2: Truncate flexible loops
Especially long loops (>20 residues)
GPCRs: ICL3 is classic target
Rule 3: Remove signal peptide
Signal peptide directs to ER
Not needed for recombinant expression
Predict with SignalP
Understanding Your Problem: Quick Diagnostic
Symptom | Likely Problem | Quick Test |
|---|---|---|
AlphaFold has long orange/red termini | Disordered termini | Check pLDDT <50 |
Internal orange/red loops (>20 residues) | Flexible loops | Check pLDDT, compare homologs |
Protein insoluble after truncation | Wrong domain boundary | Check if cut through helix/strand |
Protein inactive after expression | Removed essential domain | Check Pfam, homolog structures |
Low expression despite tag | Bad tag placement | Try opposite terminus |
Membrane protein won't insert | Tag blocks signal peptide | Remove N-terminal tag |
Key Takeaway
Construct boundary design isn't guesswork. It's a systematic engineering problem with predictable failure modes:
Disordered termini (40%): Include flexible tails that prevent crystallization
Flexible loops (20%): Internal disorder blocks structure determination
Wrong domain boundaries (30%): Remove essential structural elements
Bad tag placement (10%): Tags disrupt function or folding
Understanding your boundary problem is the first step to fixing it.

