Blog
How to Design Optimal Construct Boundaries: Step-by-Step Workflow
Jan 12, 2026
In this article, we'll show you exactly how to design optimal boundaries before you clone.
The paradigm shift: Stop guessing. Start predicting.
Traditional approach: Clone full-length → Fail → Guess truncations → Fail → Test 10-15 constructs → Maybe succeed
Modern AI-driven approach: Analyze structure → Predict boundaries → Design 2-3 rational constructs → Test → Succeed
This guide provides the complete workflow, from sequence to optimized construct.
Key Takeaways
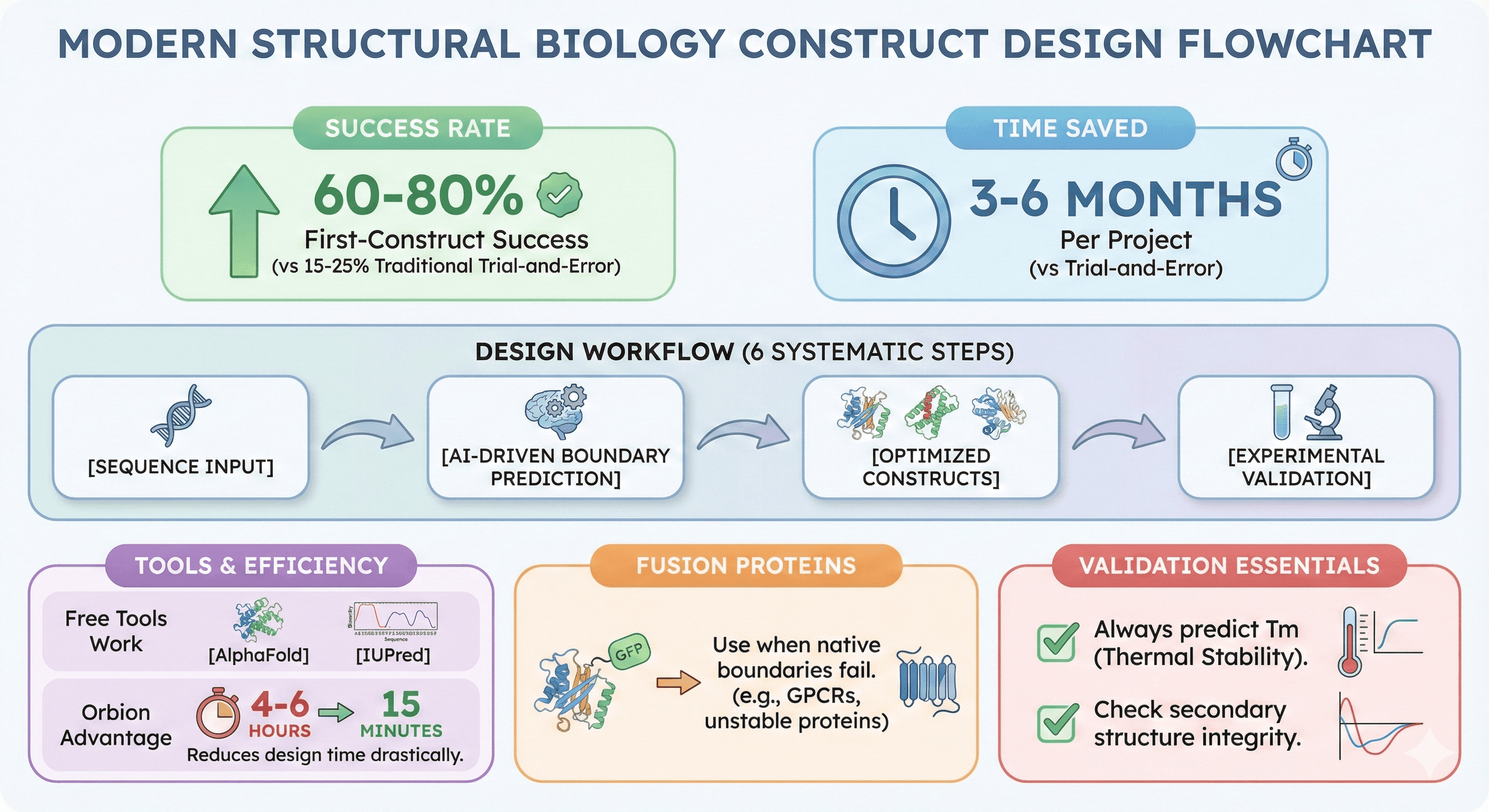
Design workflow: 6 systematic steps (sequence → boundaries → constructs → validation)
Success rate: 60-80% first-construct success with AI predictions (vs 15-25% traditional)
Time saved: 3-6 months per project (vs trial-and-error)
Tools: Free tools work (AlphaFold, IUPred), but Orbion reduces design time from 4-6 hours to 15 minutes
Fusion proteins: Use when native boundaries fail (GPCRs, unstable proteins)
Validation: Always predict Tm, check secondary structure integrity
The 6-Step Design Workflow
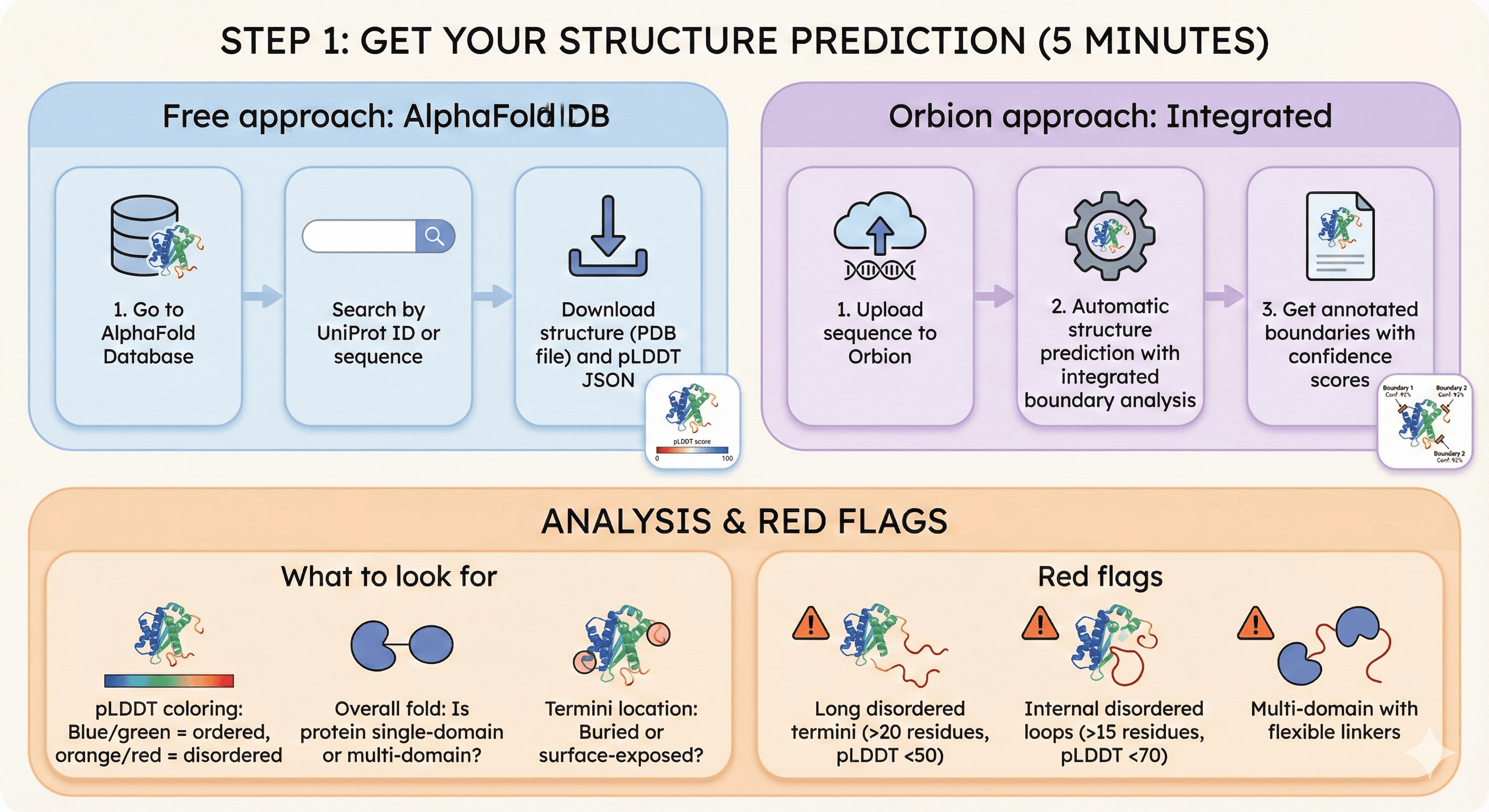
Step 1: Get Your Structure Prediction (5 minutes)
Free approach:
Go to AlphaFold Database
Search by UniProt ID or sequence
Download structure (PDB file) and pLDDT JSON
Orbion approach:
Upload sequence to Orbion
Automatic structure prediction with integrated boundary analysis
Get annotated boundaries with confidence scores
What to look for:
pLDDT coloring: Blue/green = ordered, orange/red = disordered
Overall fold: Is protein single-domain or multi-domain?
Termini location: Buried or surface-exposed?
Red flags:
Long disordered termini (>20 residues, pLDDT <50)
Internal disordered loops (>15 residues, pLDDT <70)
Multi-domain with flexible linkers
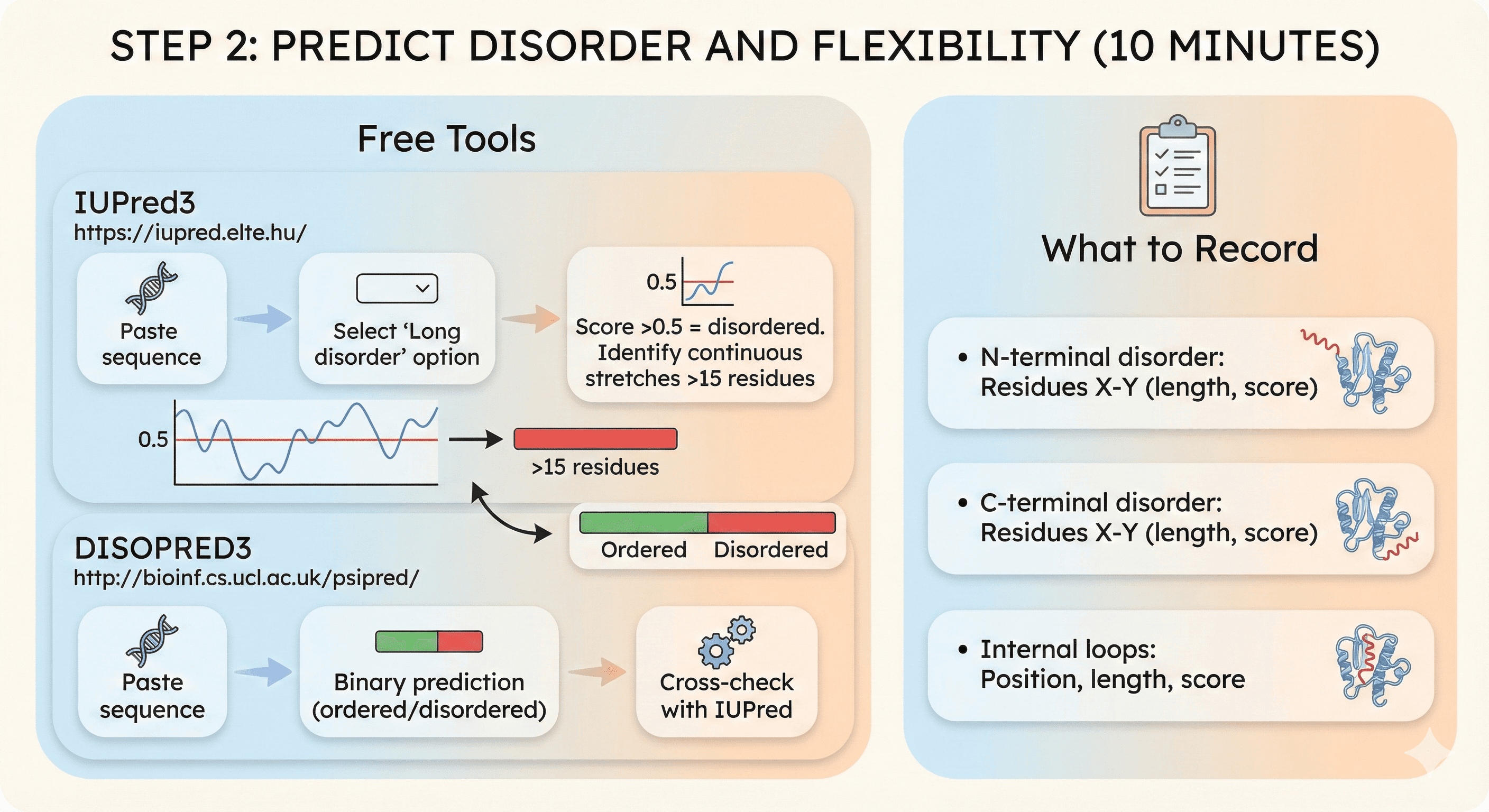
Step 2: Predict Disorder and Flexibility (10 minutes)
Free tools:
IUPred3: https://iupred.elte.hu/
Paste sequence
Select "Long disorder" option
Score >0.5 = disordered
Identify continuous stretches >15 residues
DISOPRED3: http://bioinf.cs.ucl.ac.uk/psipred/
Paste sequence
Binary prediction (ordered/disordered)
Cross-check with IUPred
What to record:
N-terminal disorder: Residues X-Y (length, score)
C-terminal disorder: Residues X-Y (length, score)
Internal loops: Position, length, score
Example output:
Step 3: Analyze Domain Architecture (10 minutes)
Free tools:
Pfam: https://pfam.xfam.org/
Paste sequence
Identifies conserved domains
Shows boundaries for each domain
InterPro: https://www.ebi.ac.uk/interpro/
Comprehensive domain annotation
Includes functional sites
Critical rules:
Don't truncate within a Pfam domain (unless you know what you're doing)
Keep complete secondary structure elements (don't cut helices/strands)
Truncate in loops between domains or elements
Example:

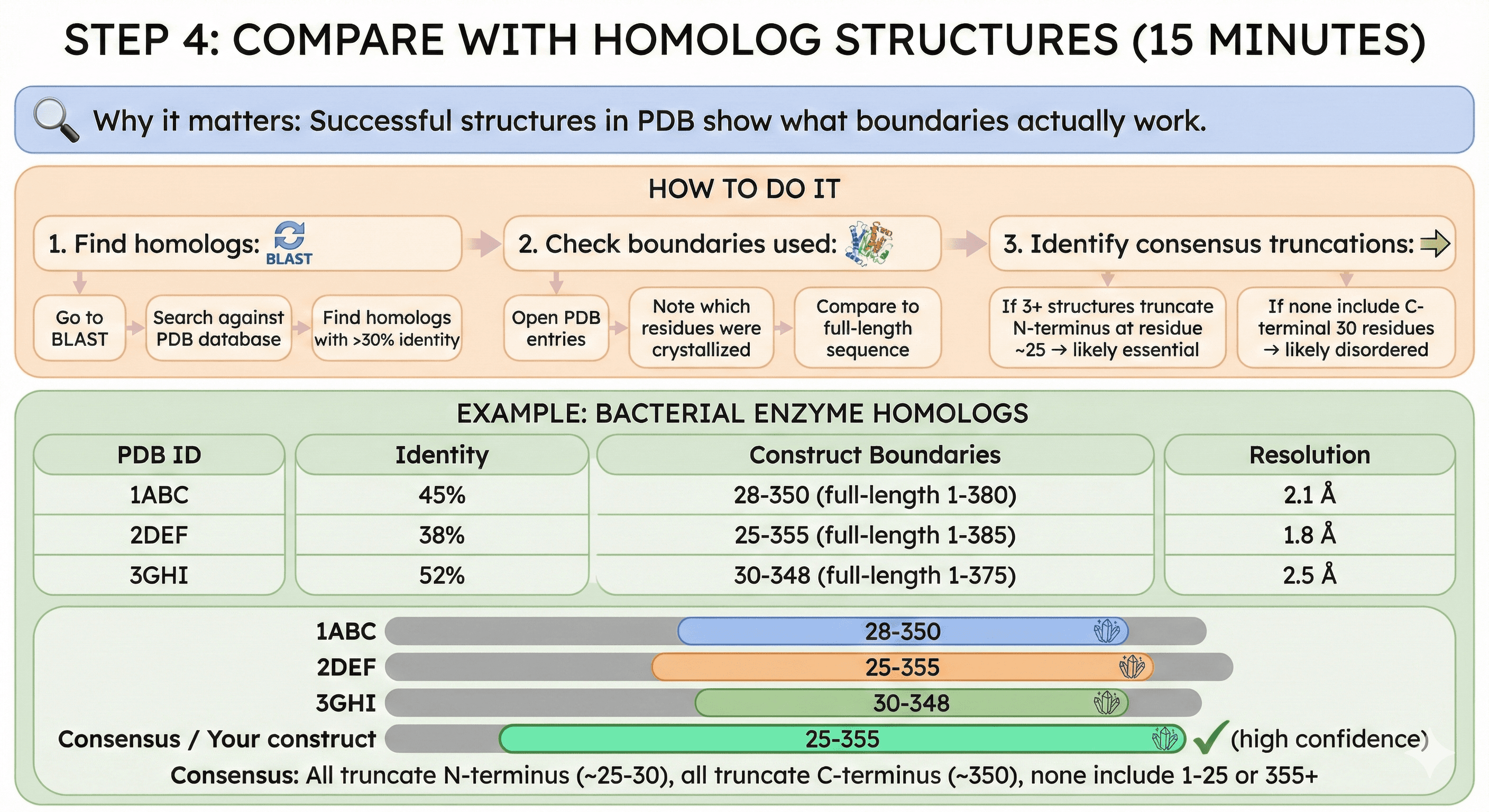
Step 4: Compare with Homolog Structures (15 minutes)
Why it matters: Successful structures in PDB show what boundaries actually work.
How to do it:
1. Find homologs:
Go to BLAST
Search against PDB database
Find homologs with >30% identity
2. Check boundaries used:
Open PDB entries
Note which residues were crystallized
Compare to full-length sequence
3. Identify consensus truncations:
If 3+ structures truncate N-terminus at residue ~25 → likely essential
If none include C-terminal 30 residues → likely disordered
Example: Bacterial enzyme homologs
PDB ID | Identity | Construct Boundaries | Resolution |
|---|---|---|---|
1ABC | 45% | 28-350 (full-length 1-380) | 2.1 Å |
2DEF | 38% | 25-355 (full-length 1-385) | 1.8 Å |
3GHI | 52% | 30-348 (full-length 1-375) | 2.5 Å |
Consensus: All truncate N-terminus (~25-30), all truncate C-terminus (~350), none include 1-25 or 355+
Your construct: 25-355 (high confidence)
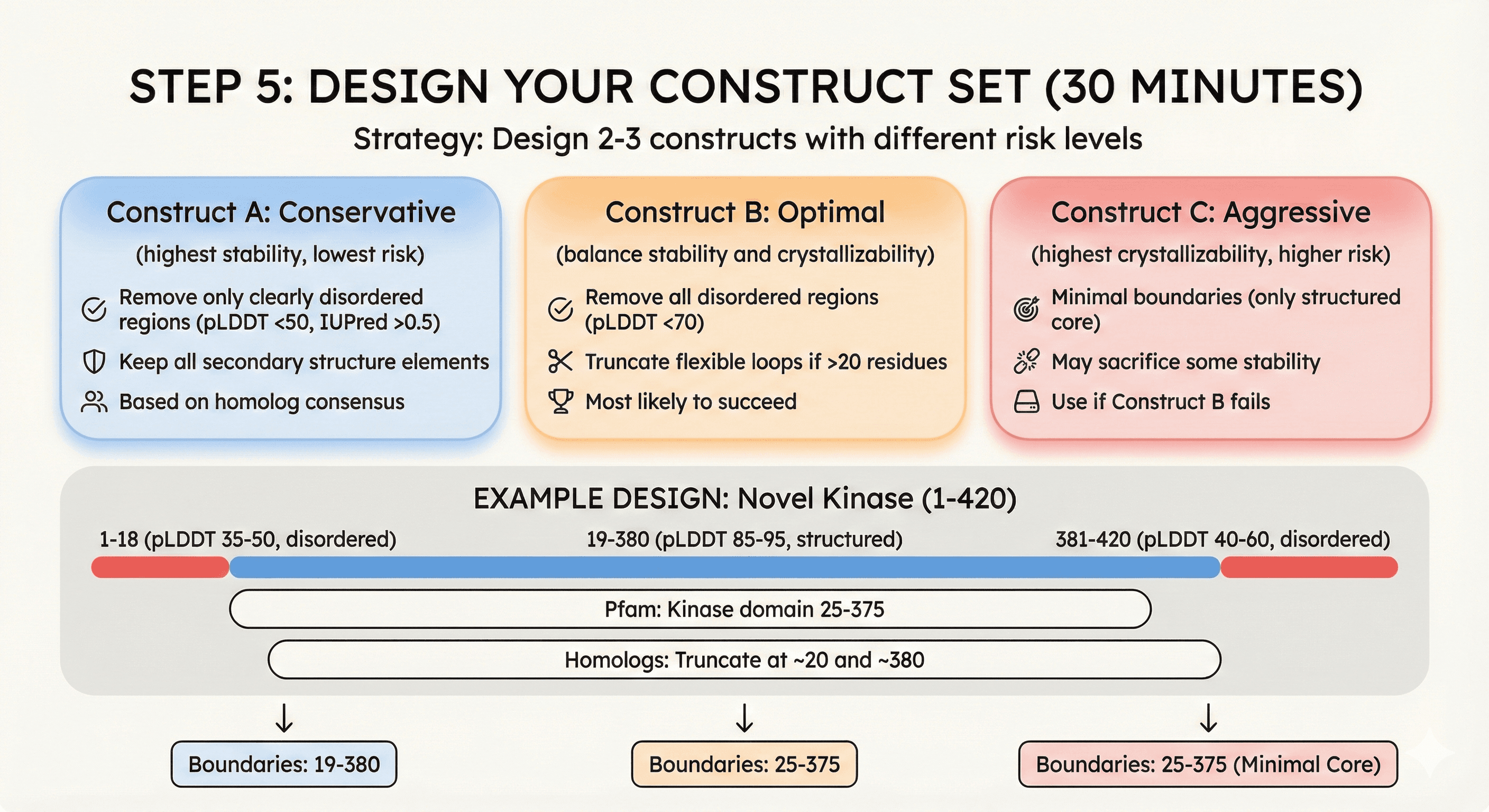
Step 5: Design Your Construct Set (30 minutes)
Strategy: Design 2-3 constructs with different risk levels
Construct A: Conservative (highest stability, lowest risk)
Remove only clearly disordered regions (pLDDT <50, IUPred >0.5)
Keep all secondary structure elements
Based on homolog consensus
Construct B: Optimal (balance stability and crystallizability)
Remove all disordered regions (pLDDT <70)
Truncate flexible loops if >20 residues
Most likely to succeed
Construct C: Aggressive (highest crystallizability, higher risk)
Minimal boundaries (only structured core)
May sacrifice some stability
Use if Construct B fails
Example design:
Target: Novel kinase (1-420)
AlphaFold pLDDT:
1-18: pLDDT 35-50 (disordered)
19-380: pLDDT 85-95 (structured)
381-420: pLDDT 40-60 (disordered)
Pfam: Kinase domain 25-375
Homologs: Truncate at ~20 and ~380
Construct set:
Which to test first: Construct B (optimal balance)
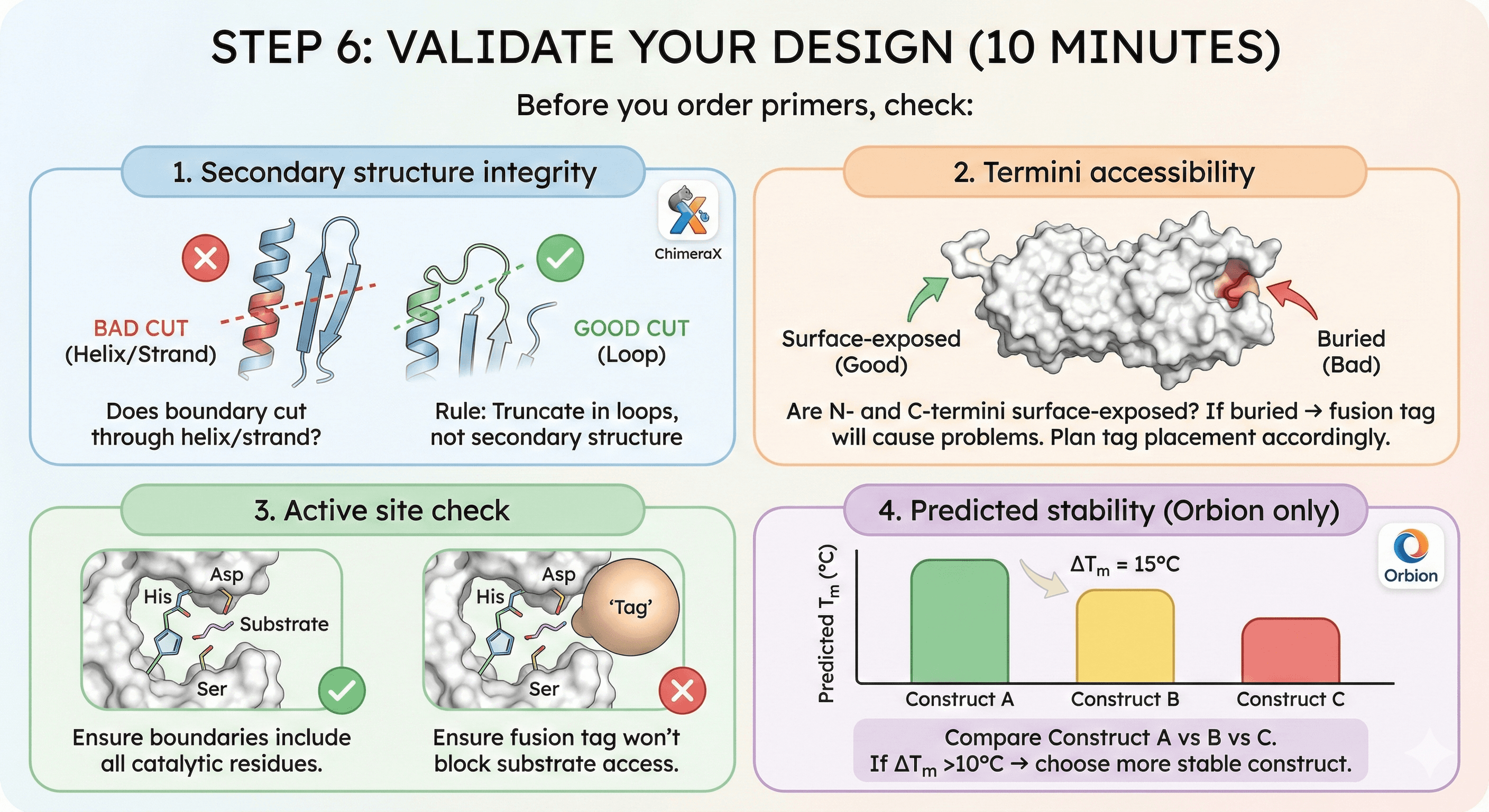
Step 6: Validate Your Design (10 minutes)
Before you order primers, check:
1. Secondary structure integrity:
Load AlphaFold structure in PyMOL or ChimeraX
Check N-terminal boundary: Does it cut through helix/strand?
Check C-terminal boundary: Same check
Rule: Truncate in loops, not secondary structure
2. Termini accessibility:
Are N- and C-termini surface-exposed?
If buried → fusion tag will cause problems
Plan tag placement accordingly
3. Active site check:
Visualize active site (if known)
Ensure your boundaries include all catalytic residues
Ensure fusion tag won't block substrate access
4. Predicted stability (Orbion only):
Orbion predicts Tm for each construct
Compare Construct A vs B vs C
If ΔTm >10°C → choose more stable construct
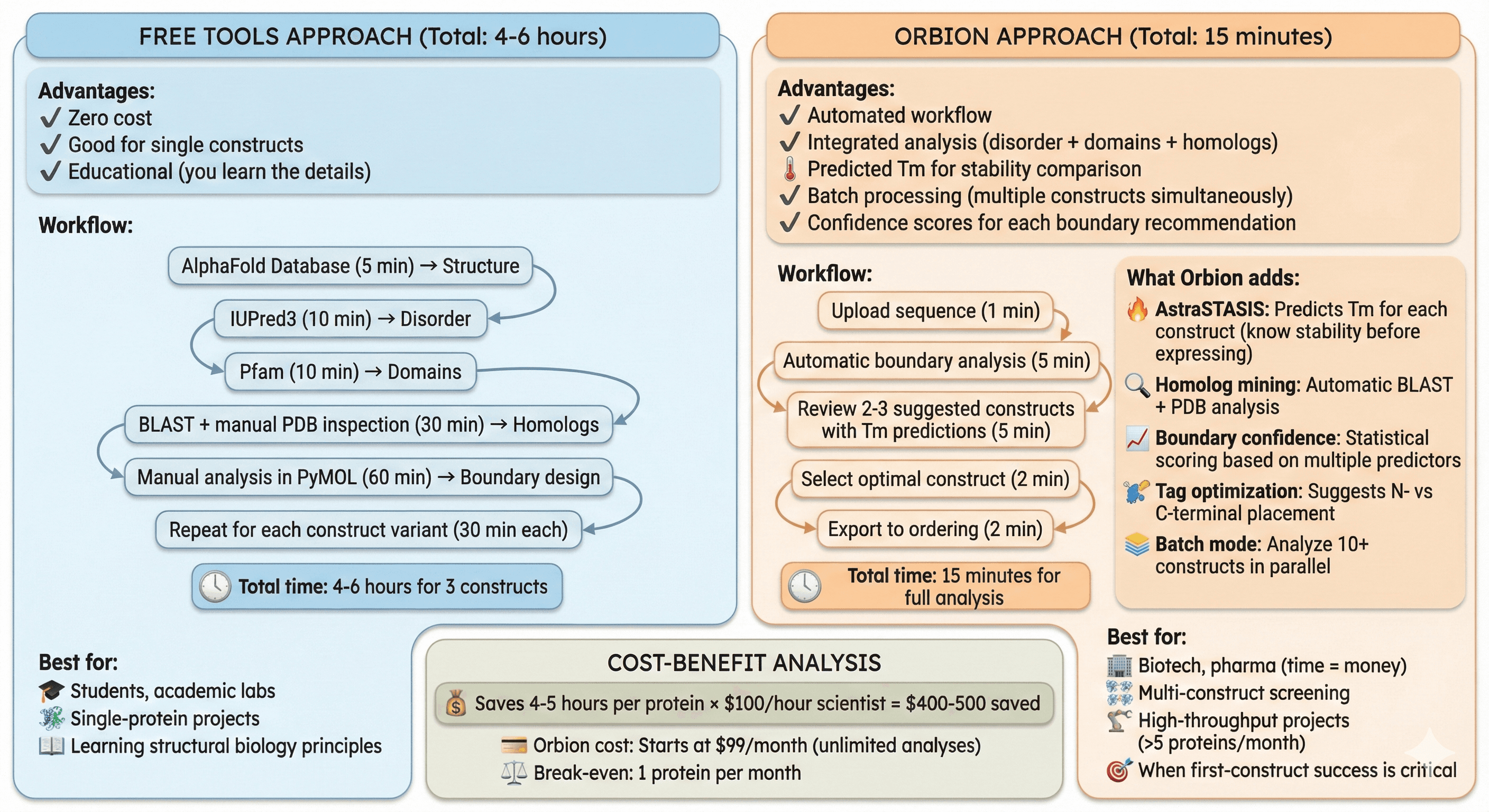
Free Tools vs Orbion: When to Use What
Free Tools Approach (Total: 4-6 hours)
Advantages:
Zero cost
Good for single constructs
Educational (you learn the details)
Workflow:
AlphaFold Database (5 min) → Structure
IUPred3 (10 min) → Disorder
Pfam (10 min) → Domains
BLAST + manual PDB inspection (30 min) → Homologs
Manual analysis in PyMOL (60 min) → Boundary design
Repeat for each construct variant (30 min each)
Total time: 4-6 hours for 3 constructs
Best for:
Students, academic labs
Single-protein projects
Learning structural biology principles
Orbion Approach (Total: 15 minutes)
Advantages:
Automated workflow
Integrated analysis (disorder + domains + homologs)
Predicted Tm for stability comparison
Batch processing (multiple constructs simultaneously)
Confidence scores for each boundary recommendation
Workflow:
Upload sequence (1 min)
Automatic boundary analysis (5 min)
Review 2-3 suggested constructs with Tm predictions (5 min)
Select optimal construct (2 min)
Export to ordering (2 min)
Total time: 15 minutes for full analysis
What Orbion adds:
AstraSTASIS: Predicts Tm for each construct (know stability before expressing)
Homolog mining: Automatic BLAST + PDB analysis
Boundary confidence: Statistical scoring based on multiple predictors
Tag optimization: Suggests N- vs C-terminal placement
Batch mode: Analyze 10+ constructs in parallel
Best for:
Biotech, pharma (time = money)
Multi-construct screening
High-throughput projects (>5 proteins/month)
When first-construct success is critical
Cost-benefit:
Saves 4-5 hours per protein × $100/hour scientist = $400-500 saved
Orbion cost: Starts at $99/month (unlimited analyses)
Break-even: 1 protein per month
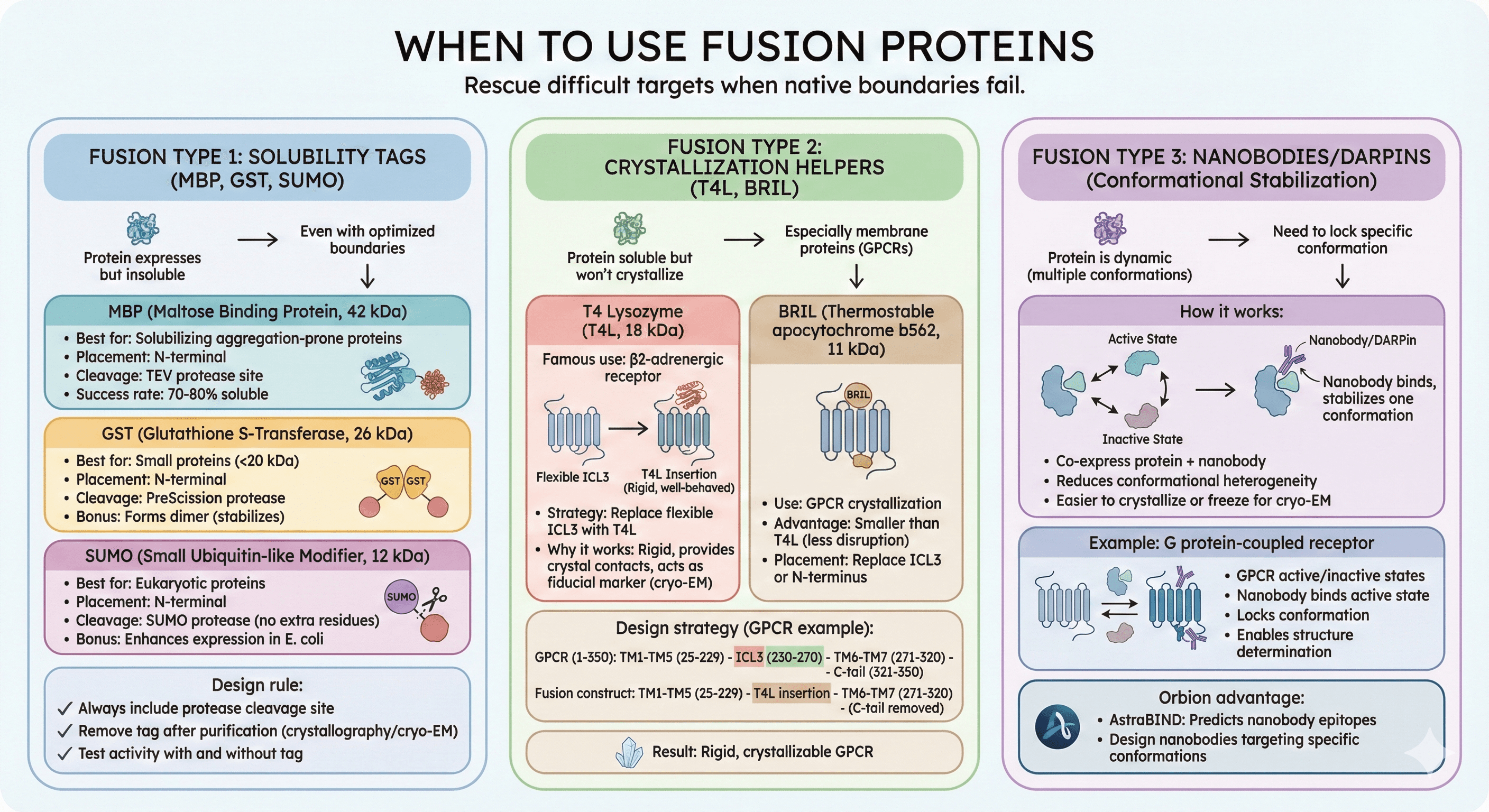
When to Use Fusion Proteins
Sometimes native boundaries aren't enough. Fusion proteins can rescue difficult targets.
Fusion Type 1: Solubility Tags (MBP, GST, SUMO)
When to use:
Protein expresses but insoluble
Even with optimized boundaries
Common tags:
MBP (Maltose Binding Protein, 42 kDa):
Best for: Solubilizing aggregation-prone proteins
Placement: N-terminal
Cleavage: TEV protease site
Success rate: 70-80% of insoluble proteins become soluble
GST (Glutathione S-Transferase, 26 kDa):
Best for: Small proteins (<20 kDa)
Placement: N-terminal
Cleavage: PreScission protease
Bonus: Forms dimer (stabilizes small proteins)
SUMO (Small Ubiquitin-like Modifier, 12 kDa):
Best for: Eukaryotic proteins
Placement: N-terminal
Cleavage: SUMO protease (leaves no extra residues)
Bonus: Enhances expression in E. coli
Design rule:
Always include protease cleavage site
Remove tag after purification for crystallography/cryo-EM
Test activity with and without tag
Fusion Type 2: Crystallization Helpers (T4L, BRIL)
When to use:
Protein soluble but won't crystallize
Especially for membrane proteins (GPCRs)
T4 Lysozyme (T4L, 18 kDa):
Famous use: β2-adrenergic receptor (2007 Nobel Prize)
Strategy: Replace flexible ICL3 with T4L
Why it works:
Rigid, well-behaved protein
Provides crystal contacts
Acts as fiducial marker for cryo-EM alignment
BRIL (Thermostable apocytochrome b562, 11 kDa):
Use: GPCR crystallization
Advantage: Smaller than T4L (less disruption)
Placement: Replace ICL3 or insert at N-terminus
Design strategy:
Fusion Type 3: Nanobodies/DARPins (Conformational Stabilization)
When to use:
Protein is dynamic (multiple conformations)
Need to lock specific conformation for structure
How it works:
Co-express protein + nanobody
Nanobody binds, stabilizes one conformation
Reduces conformational heterogeneity
Easier to crystallize or freeze for cryo-EM
Example: G protein-coupled receptor
GPCR has active and inactive states
Nanobody binds active state
Locks conformation
Enables structure determination of active state
Orbion advantage:
AstraBIND: Predicts nanobody epitopes
Design nanobodies targeting specific conformations
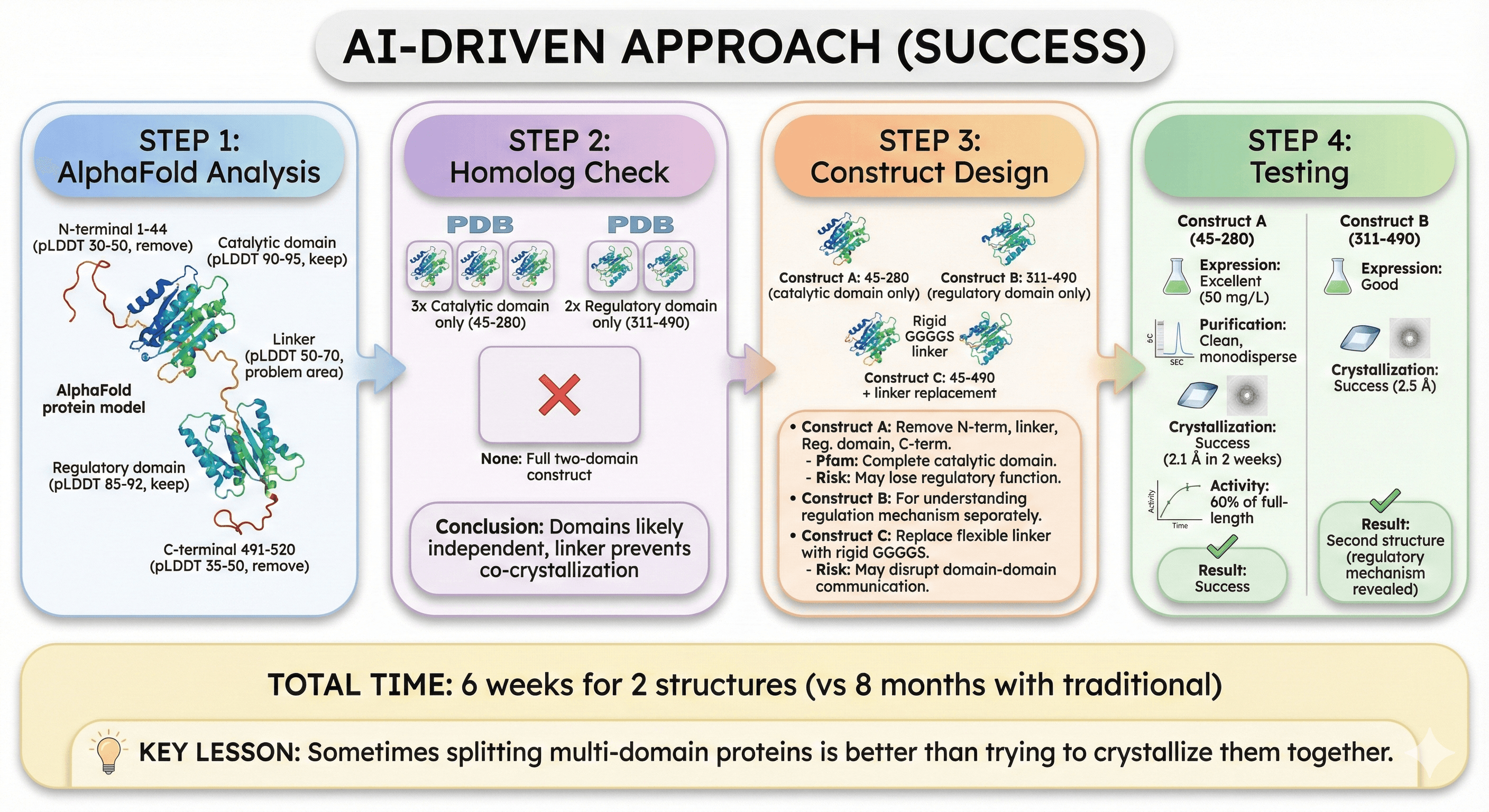
Case Study: Multi-Domain Protein Optimization
Target: Novel bacterial enzyme with regulatory domain
Full-length: 1-520
N-terminal tail: 1-44 (disordered)
Catalytic domain: 45-280 (structured)
Linker: 281-310 (flexible)
Regulatory domain: 311-490 (structured)
C-terminal tail: 491-520 (disordered)
Traditional Approach (Failed)
Attempt 1: Full-length (1-520)
Expression: Low yield
Purification: Multiple degradation bands
Crystallization: No crystals after 300 conditions
Result: Failure (disordered tails cause problems)
Attempt 2: Remove tails (45-490)
Expression: Good
Purification: Clean, but some degradation
Crystallization: No crystals
Analysis: Limited proteolysis shows cleavage in linker (281-310)
Result: Partial success (linker still flexible)
Total time: 8 months, 2 failed attempts
AI-Driven Approach (Success)
Step 1: AlphaFold analysis
N-terminal 1-44: pLDDT 30-50 (remove)
Catalytic domain: pLDDT 90-95 (keep)
Linker: pLDDT 50-70 (problem area)
Regulatory domain: pLDDT 85-92 (keep)
C-terminal 491-520: pLDDT 35-50 (remove)
Step 2: Homolog check
Found 5 homologs in PDB
3 structures: Catalytic domain only (45-280)
2 structures: Regulatory domain only (311-490)
None: Full two-domain construct
Conclusion: Domains likely independent, linker prevents co-crystallization
Step 3: Construct design
Step 4: Testing
Construct A (45-280):
Expression: Excellent (50 mg/L)
Purification: Clean, monodisperse
Crystallization: Success (2.1 Å structure in 2 weeks)
Activity: 60% of full-length (good enough for structural studies)
Result: Success
Construct B (311-490):
Expression: Good
Crystallization: Success (2.5 Å)
Result: Second structure (regulatory mechanism revealed)
Total time: 6 weeks for 2 structures (vs 8 months with traditional)
Key lesson: Sometimes splitting multi-domain proteins is better than trying to crystallize them together.
Membrane Proteins: Special Workflow
Membrane proteins require additional considerations.
Extra Steps for Membrane Proteins
1. Predict transmembrane helices:
DeepTMHMM: https://dtu.biolib.com/DeepTMHMM
Predicts TM helix positions
Identifies inside/outside orientation
Output:
2. Define TM boundaries:
Rules:
Include complete TM helices (don't truncate in middle)
Start boundary 5-10 residues before first TM helix
End boundary 5-10 residues after last TM helix
Remove signal peptide (not needed for recombinant expression)
Truncate long intracellular/extracellular loops
3. Handle flexible loops:
GPCRs: Intracellular loop 3 (ICL3) is usually 20-80 residues, highly flexible
Options:
Option A: Remove ICL3 entirely (if not functionally critical)
Option B: Replace with short linker (GGGGS)
Option C: Replace with T4 lysozyme (gold standard for crystallization)
Example construct:
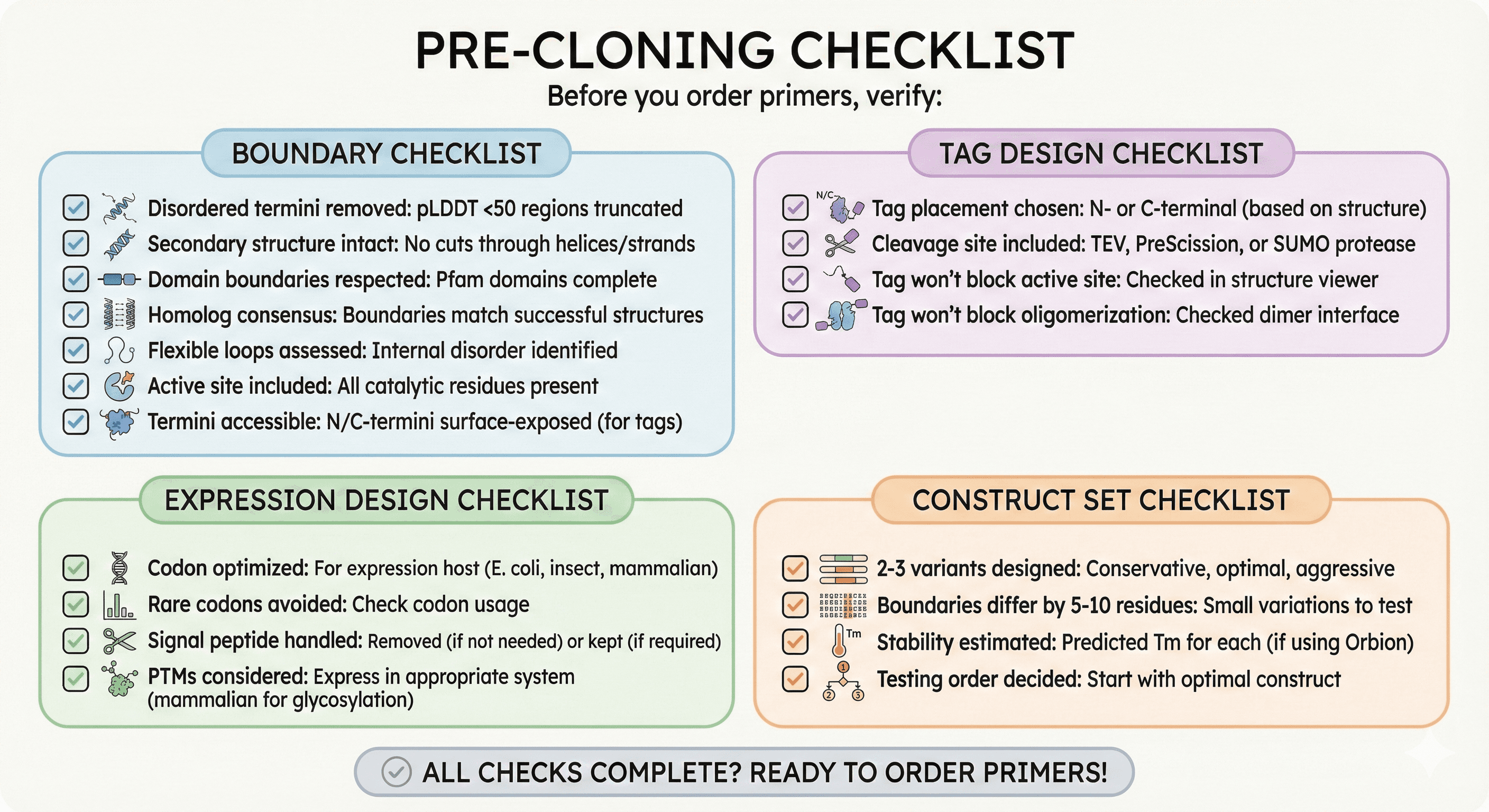
Pre-Cloning Checklist
Before you order primers, verify:
Boundary Checklist
[ ] Disordered termini removed: pLDDT <50 regions truncated
[ ] Secondary structure intact: No cuts through helices/strands
[ ] Domain boundaries respected: Pfam domains complete
[ ] Homolog consensus: Boundaries match successful structures
[ ] Flexible loops assessed: Internal disorder identified
[ ] Active site included: All catalytic residues present
[ ] Termini accessible: N/C-termini surface-exposed (for tags)
Tag Design Checklist
[ ] Tag placement chosen: N- or C-terminal (based on structure)
[ ] Cleavage site included: TEV, PreScission, or SUMO protease
[ ] Tag won't block active site: Checked in structure viewer
[ ] Tag won't block oligomerization: Checked dimer interface
Expression Design Checklist
[ ] Codon optimized: For expression host (E. coli, insect, mammalian)
[ ] Rare codons avoided: Check codon usage
[ ] Signal peptide handled: Removed (if not needed) or kept (if required)
[ ] PTMs considered: Express in appropriate system (mammalian for glycosylation)
Construct Set Checklist
[ ] 2-3 variants designed: Conservative, optimal, aggressive
[ ] Boundaries differ by 5-10 residues: Small variations to test
[ ] Stability estimated: Predicted Tm for each (if using Orbion)
[ ] Testing order decided: Start with optimal construct
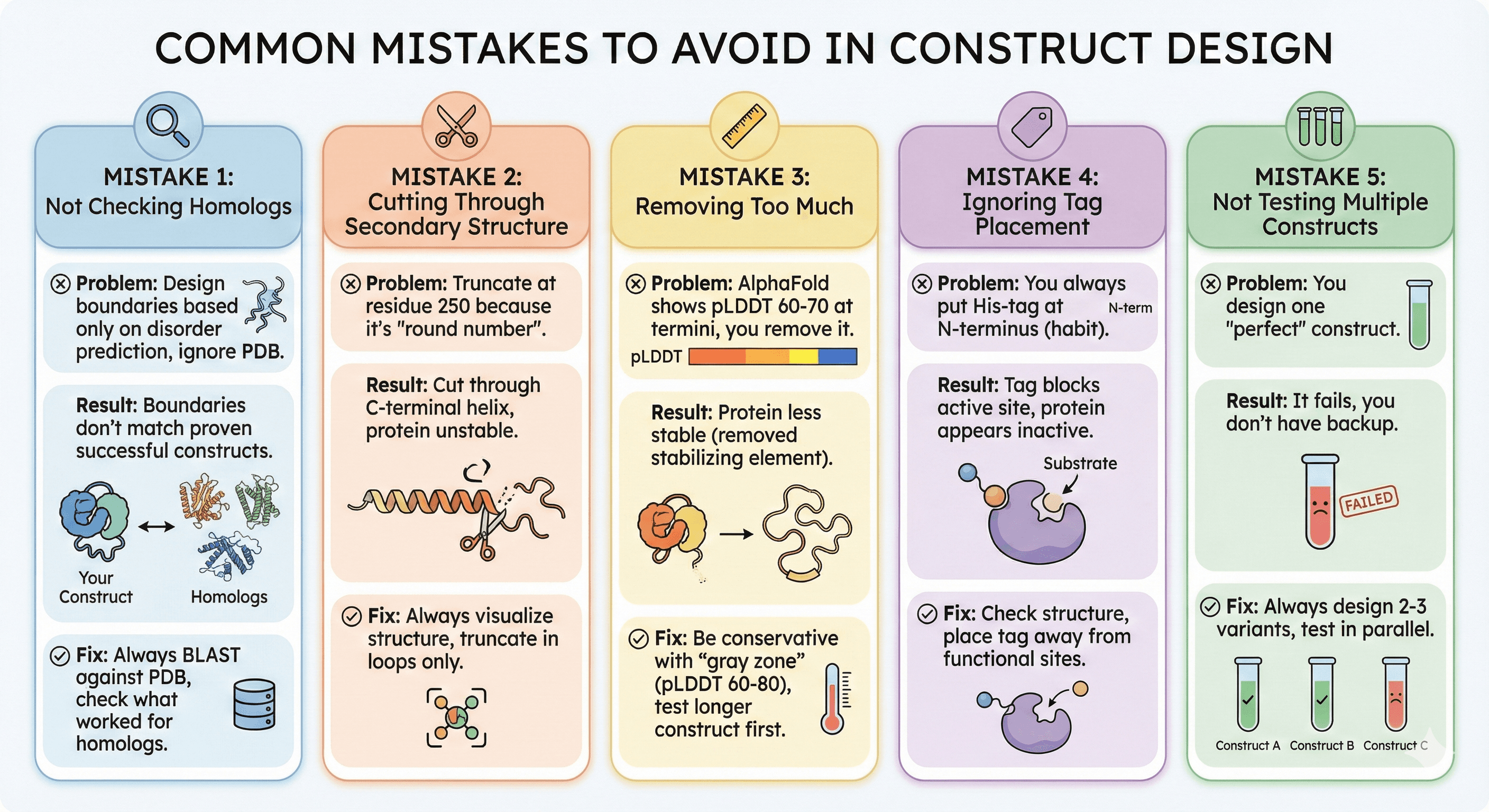
Common Mistakes to Avoid
Mistake 1: Not Checking Homologs
Problem: You design boundaries based only on disorder prediction, ignore PDB
Result: Your boundaries don't match proven successful constructs
Fix: Always BLAST against PDB, check what worked for homologs
Mistake 2: Cutting Through Secondary Structure
Problem: You truncate at residue 250 because it's "round number"
Result: You cut through C-terminal helix, protein unstable
Fix: Always visualize structure, truncate in loops only
Mistake 3: Removing Too Much
Problem: AlphaFold shows pLDDT 60-70 at termini, you remove it
Result: Protein less stable (removed stabilizing element)
Fix: Be conservative with "gray zone" (pLDDT 60-80), test longer construct first
Mistake 4: Ignoring Tag Placement
Problem: You always put His-tag at N-terminus (habit)
Result: Tag blocks active site, protein appears inactive
Fix: Check structure, place tag away from functional sites
Mistake 5: Not Testing Multiple Constructs
Problem: You design one "perfect" construct
Result: It fails, you don't have backup
Fix: Always design 2-3 variants, test in parallel
Advanced: When Standard Boundaries Fail
Problem: All Constructs Aggregate
You've tried:
Optimal boundaries (removed disorder)
Conservative boundaries (kept more)
Aggressive boundaries (minimal)
All aggregate during expression or purification
Solutions:
1. Try fusion tags (MBP, GST, SUMO):
MBP most effective for aggregation
Express as MBP-fusion, cleave after purification
2. Change expression system:
If E. coli fails → Try insect cells (Sf9/High Five)
If insect fails → Try mammalian (HEK293, CHO)
3. Lower expression temperature:
Express at 18°C instead of 37°C
Slower folding, less aggregation
4. Add chaperone co-expression:
GroEL/GroES for E. coli
Helps proper folding
5. Screen for stabilizing mutations (Orbion):
AstraSTASIS predicts stabilizing point mutations
Increase Tm by 5-15°C
More stable = less aggregation
Problem: Protein Soluble But Inactive
You've truncated boundaries, protein expresses and purifies, but has no activity
Diagnosis:
1. Check what you removed:
Did you remove part of active site?
Did you remove regulatory domain needed for activity?
Did you remove cofactor binding site?
2. Compare with full-length:
Express longer construct (more conservative boundaries)
Test activity
If active → you removed something essential
Solutions:
1. Extend boundaries:
Add back 10-20 residues at each terminus
Test which end matters
2. Include regulatory domain:
If multi-domain, include both catalytic + regulatory
Accept that you may need fusion approach for crystallization
3. Add back cofactor:
Some proteins need metal ions (Zn²⁺, Mg²⁺) or organic cofactors
Supplement during expression/purification
Key Takeaway
Construct boundary design is systematic engineering, not guesswork:
The 6-step workflow:
Get structure prediction (AlphaFold)
Predict disorder (IUPred, DISOPRED)
Analyze domains (Pfam, InterPro)
Compare homologs (BLAST + PDB)
Design 2-3 constructs (conservative/optimal/aggressive)
Validate before cloning (check secondary structure, termini)
Tools:
Free: 4-6 hours, good for learning
Orbion: 15 minutes, integrated workflow, Tm prediction
When to use fusions:
Solubility issues → MBP, GST, SUMO
Crystallization issues → T4L, BRIL
Conformational heterogeneity → Nanobodies
Success rate:
Traditional (trial-and-error): 15-25% first-construct success
AI-driven (predicted boundaries): 60-80% first-construct success
The paradigm shift is here. Stop guessing. Start predicting.

