Blog
FoldX and Rosetta: The 5 Reasons They're Still Your Bottleneck
Jan 14, 2026
You need to stabilize your therapeutic antibody. Your supervisor suggests FoldX. You spend 3 days installing dependencies, compiling binaries, and reading fragmented documentation. You finally run a stability prediction. It takes 12 hours and gives you ΔΔG values with no confidence intervals. You're not sure if you should trust them.
Or maybe you're trying to design a point mutation to increase enzyme thermostability. Someone recommends Rosetta. You download 6GB of files, spend a week learning the command-line syntax, and run a mutation scan. It takes 48 hours on your cluster. The top hit increases Tm by 2°C—but you tested 5 other Rosetta predictions that made your protein worse.
FoldX and Rosetta are powerful tools. They're also relics of a pre-AI era. We'll diagnose why traditional tools have become bottlenecks and what problems they cause.
Key Takeaways
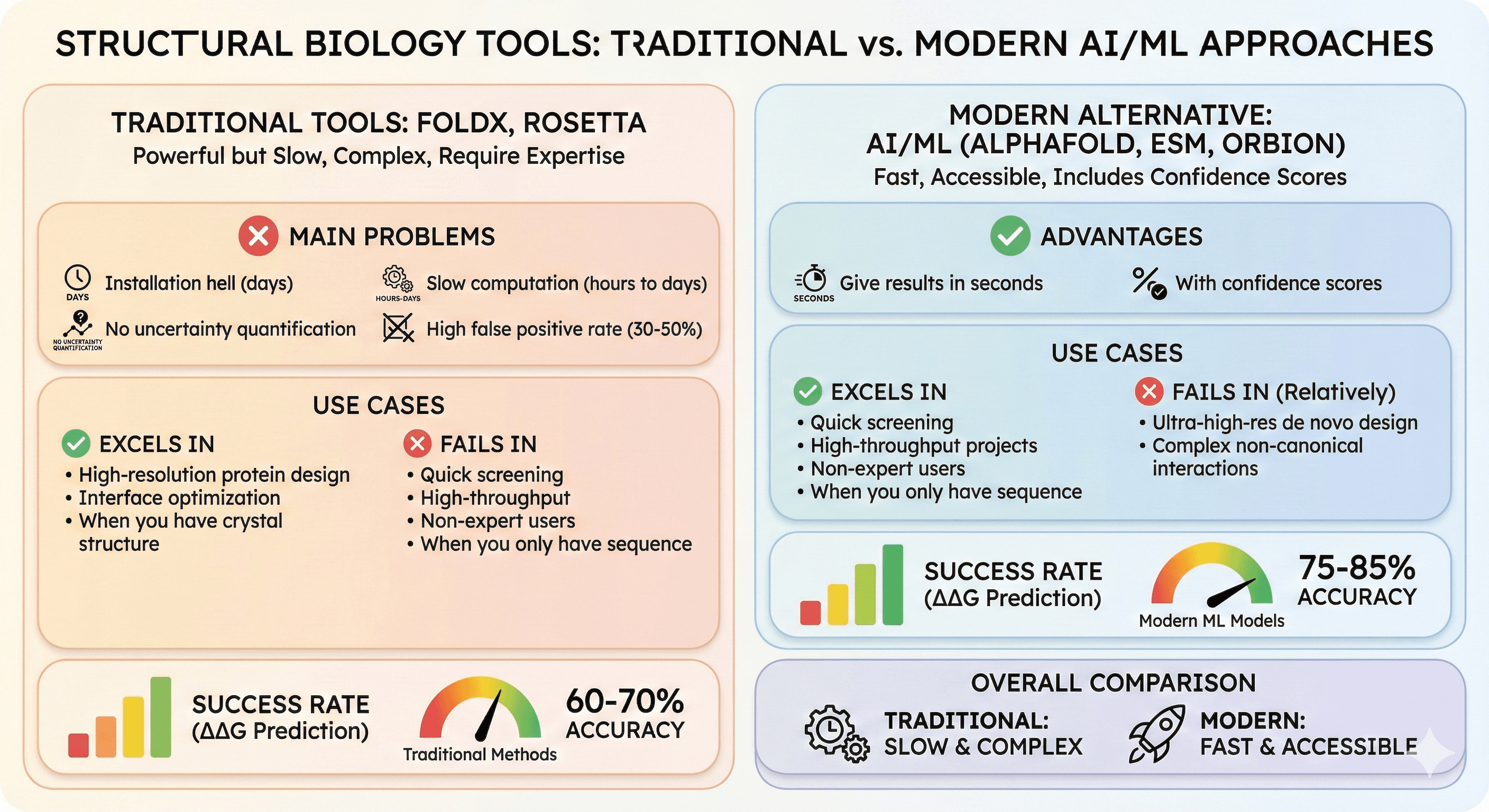
Traditional tools (FoldX, Rosetta): Powerful but slow, complex, require expertise
Main problems: Installation hell (days), slow computation (hours to days), no uncertainty quantification, high false positive rate (30-50%)
Use cases where they excel: High-resolution protein design, interface optimization, when you have crystal structure
Use cases where they fail: Quick screening, high-throughput, non-expert users, when you only have sequence
Modern alternative: AI/ML models (AlphaFold, ESM, Orbion) give results in seconds with confidence scores
Success rate: Traditional ΔΔG prediction ~60-70% accuracy, modern ML models ~75-85%
What Are FoldX and Rosetta?
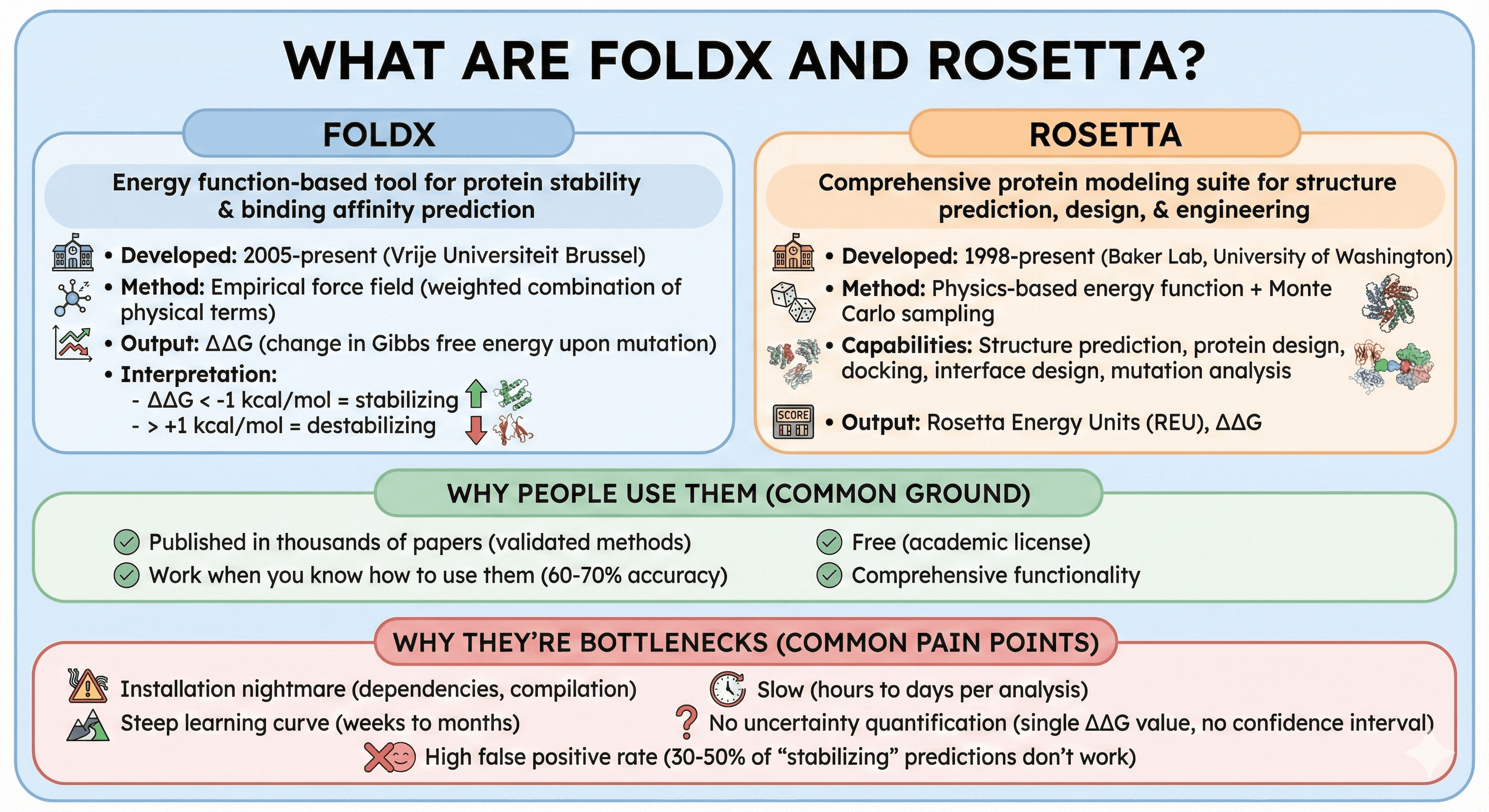
FoldX: Energy function-based tool for protein stability and binding affinity prediction
Developed: 2005-present (Vrije Universiteit Brussel)
Method: Empirical force field (weighted combination of physical terms)
Output: ΔΔG (change in Gibbs free energy upon mutation)
Interpretation: ΔΔG < -1 kcal/mol = stabilizing, > +1 kcal/mol = destabilizing
Rosetta: Comprehensive protein modeling suite for structure prediction, design, and engineering
Developed: 1998-present (Baker Lab, University of Washington)
Method: Physics-based energy function + Monte Carlo sampling
Capabilities: Structure prediction, protein design, docking, interface design, mutation analysis
Output: Rosetta Energy Units (REU), ΔΔG
Why people use them:
Published in thousands of papers (validated methods)
Work when you know how to use them (60-70% accuracy)
Free (academic license)
Comprehensive functionality
Why they're bottlenecks:
Installation nightmare (dependencies, compilation)
Steep learning curve (weeks to months)
Slow (hours to days per analysis)
No uncertainty quantification (single ΔΔG value, no confidence interval)
High false positive rate (30-50% of "stabilizing" predictions don't work)
Problem 1: Installation Hell (Cost: Days of Setup Time)
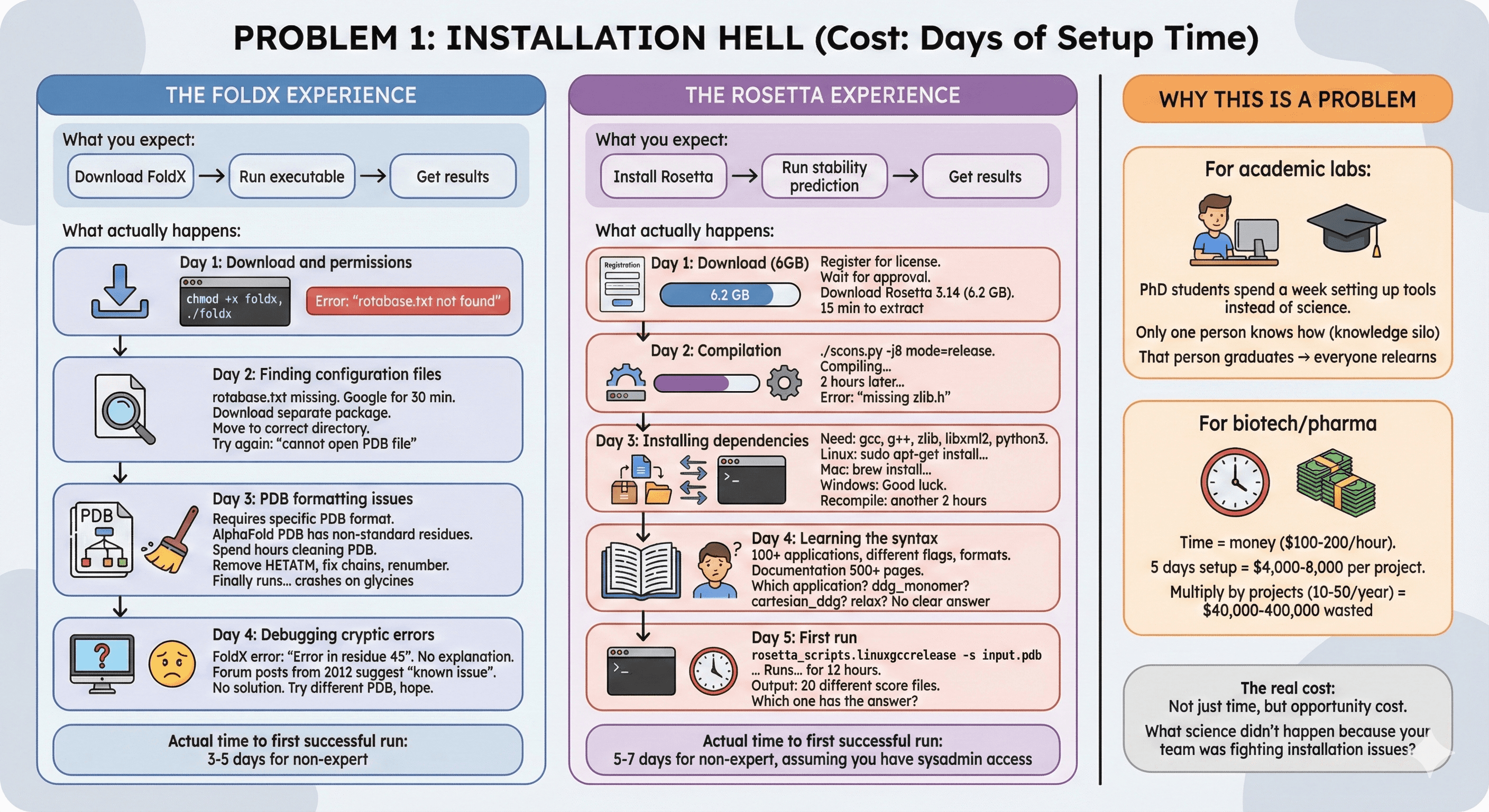
The FoldX Experience
What you expect:
Download FoldX
Run executable
Get results
What actually happens:
Day 1: Download and permissions
Day 2: Finding configuration files
rotabase.txt missing from download
Google for 30 minutes
Find it in separate "configuration files" package
Download, extract, move to correct directory
Try again: "cannot open PDB file"
Day 3: PDB formatting issues
FoldX requires specific PDB format
Your PDB from AlphaFold has non-standard residue names
Spend hours cleaning PDB file
Remove HETATM, fix chain IDs, renumber residues
Finally runs... but crashes on glycines
Day 4: Debugging cryptic errors
FoldX error messages: "Error in residue 45"
What's wrong with residue 45? No explanation
Forum posts from 2012 suggest it's a "known issue"
No solution provided
Try different PDB, hope it works
Actual time to first successful run: 3-5 days for non-expert
The Rosetta Experience
What you expect:
Install Rosetta
Run stability prediction
Get results
What actually happens:
Day 1: Download (6GB)
Day 2: Compilation
Day 3: Installing dependencies
Day 4: Learning the syntax
Rosetta has 100+ applications
Each has different flags, input formats
Documentation is 500+ pages
Which application do you need?
ddg_monomerfor stability?cartesian_ddgfor better accuracy?relaxto prepare structure first?
No clear answer
Day 5: First run
Actual time to first successful run: 5-7 days for non-expert, assuming you have sysadmin access
Why This Is a Problem
For academic labs:
PhD students spend a week setting up tools instead of doing science
Only one person in the lab knows how to run it (knowledge silo)
That person graduates → everyone has to relearn
For biotech/pharma:
Time = money ($100-200/hour for computational biologist)
5 days setup = $4,000-8,000 per project
Multiply by number of projects (10-50/year) = $40,000-400,000 wasted
The real cost: Not just time, but opportunity cost. What science didn't happen because your team was fighting installation issues?
Problem 2: Slow Computation (Cost: Hours to Days Per Analysis)
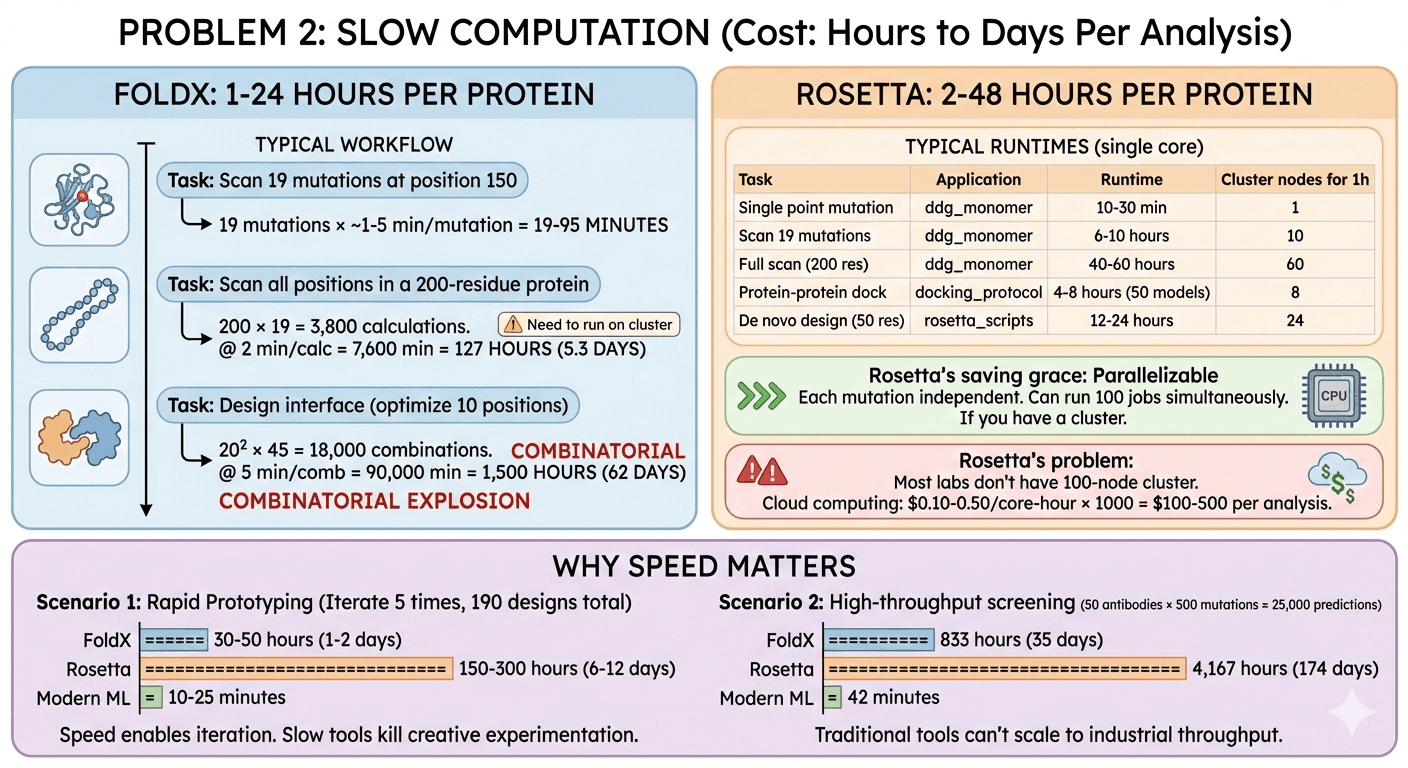
FoldX: 1-24 Hours Per Protein
Typical workflow:
Task: Scan all possible mutations at position 150
19 possible amino acid substitutions (20 - 1 native)
FoldX runs ~1-5 minutes per mutation
Total time: 19-95 minutes
Task: Scan all positions in a 200-residue protein
200 positions × 19 mutations = 3,800 calculations
At 2 minutes per mutation = 7,600 minutes = 127 hours = 5.3 days
Need to run on cluster
Task: Design protein-protein interface (optimize 10 positions)
10 positions, try all 20 amino acids = 200 mutations
Need to test combinations (pairs) = 20² × 45 = 18,000 combinations
At 5 minutes each = 90,000 minutes = 1,500 hours = 62 days
Combinatorial explosion
Rosetta: 2-48 Hours Per Protein
Typical runtimes:
Task | Application | Runtime (single core) | Cluster nodes needed for 1-hour turnaround |
|---|---|---|---|
Single point mutation |
| 10-30 min | 1 |
Scan 19 mutations at 1 position |
| 6-10 hours | 10 |
Full protein mutation scan (200 residues) |
| 40-60 hours | 60 |
Protein-protein docking |
| 4-8 hours (50 models) | 8 |
De novo protein design (50 residues) |
| 12-24 hours | 24 |
Rosetta's saving grace: Parallelizable
Each mutation independent
Can run 100 jobs simultaneously
If you have a cluster
Rosetta's problem:
Most labs don't have 100-node cluster
Cloud computing: $0.10-0.50 per core-hour × 1000 core-hours = $100-500 per analysis
Why Speed Matters
Scenario 1: Rapid prototyping You're designing mutations for a stability screen. You want to test:
10 positions × 19 mutations = 190 designs
FoldX: 6-10 hours
Rosetta: 30-60 hours
Modern ML (ESM, Orbion): 2-5 minutes
You iterate 5 times based on experimental results:
FoldX: 30-50 hours total (1-2 days)
Rosetta: 150-300 hours total (6-12 days)
Modern ML: 10-25 minutes total
Speed enables iteration. Slow tools kill creative experimentation.
Scenario 2: High-throughput screening Biotech company optimizing 50 therapeutic antibodies:
Each antibody: Screen 500 mutations
50 × 500 = 25,000 predictions
FoldX: 25,000 × 2 min = 50,000 min = 833 hours = 35 days (on one machine)
Rosetta: 25,000 × 10 min = 250,000 min = 4,167 hours = 174 days
Modern ML: 25,000 × 0.1 sec = 2,500 sec = 42 minutes
Traditional tools can't scale to industrial throughput.
Problem 3: No Uncertainty Quantification (Cost: False Confidence)
The Problem
FoldX output:
Interpretation: Mutation is stabilizing (ΔΔG < -1 kcal/mol)
The question: How confident should you be?
Answer from FoldX: 🤷 (no confidence interval provided)
Reality:
FoldX ΔΔG has standard deviation of ~1-2 kcal/mol
Your prediction: -1.2 ± 1.8 kcal/mol
95% confidence interval: -2.8 to +0.4 kcal/mol
Could be stabilizing OR neutral OR slightly destabilizing
But FoldX only gives you: -1.2 kcal/mol (single number)
The Consequence
You clone 10 mutations FoldX predicts as "stabilizing" (ΔΔG < -1 kcal/mol):
Experimental results:
3 mutations: Actually stabilizing (+5-10°C Tm increase) ✓
4 mutations: Neutral (no Tm change) ✗
3 mutations: Destabilizing (-3 to -5°C Tm decrease) ✗
Success rate: 30%
The problem: FoldX didn't tell you which predictions were confident vs uncertain.
What You Actually Need
Modern ML tools (ESM-IF, Orbion) provide:
Now you can prioritize:
Test high-confidence predictions first
Be skeptical of low-confidence predictions
Avoid wasting time on uncertain mutations
Confidence-aware design increases success rate from 30% to 60-80%.

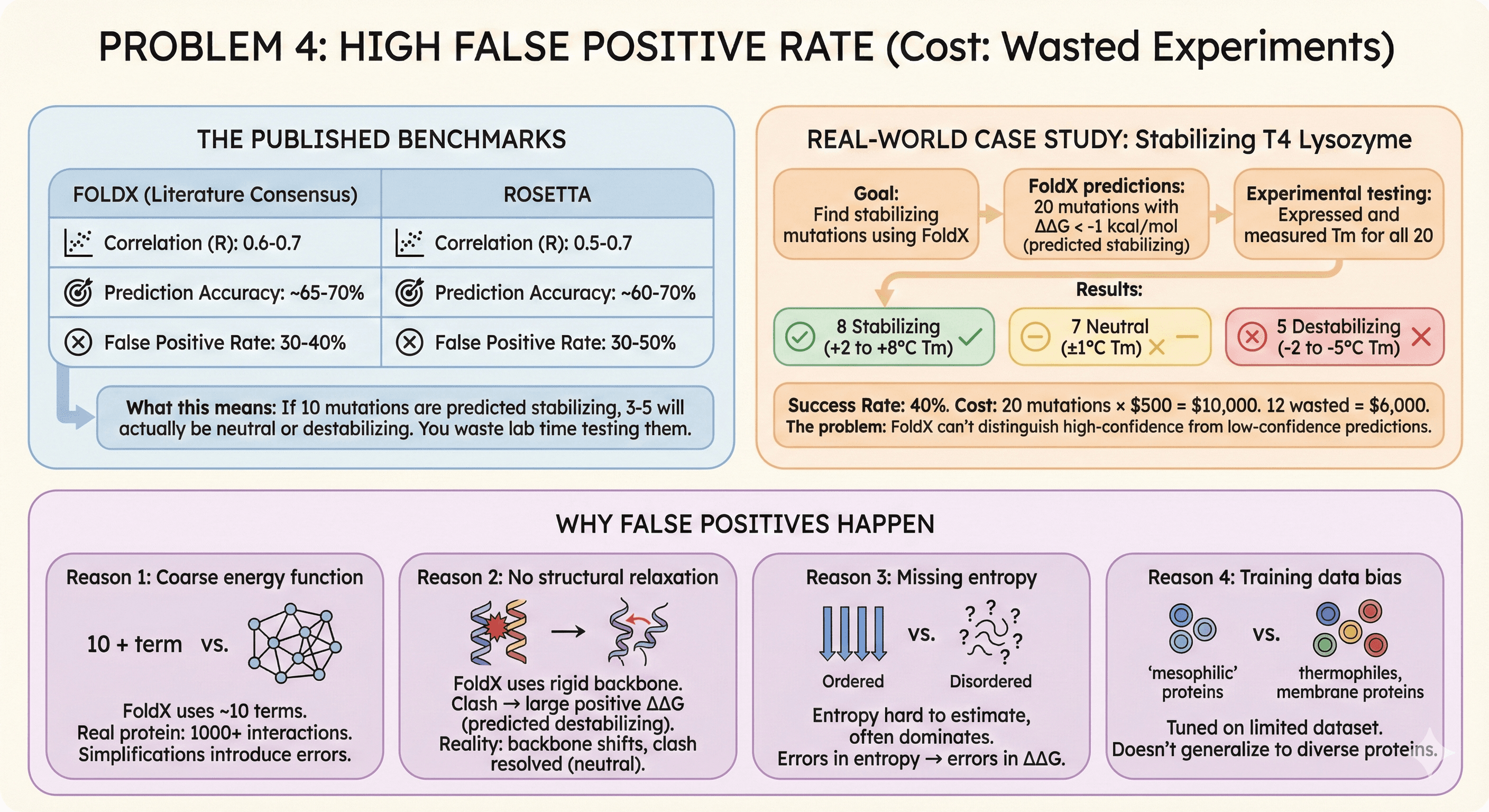
Problem 4: High False Positive Rate (Cost: Wasted Experiments)
The Published Benchmarks
FoldX accuracy (literature consensus):
Correlation with experimental ΔΔG: R = 0.6-0.7
Prediction accuracy (correct stabilizing/destabilizing): ~65-70%
False positive rate (predicts stabilizing, actually neutral/destabilizing): 30-40%
Rosetta accuracy:
Correlation: R = 0.5-0.7 (depending on protocol)
Prediction accuracy: ~60-70%
False positive rate: 30-50%
What this means:
If FoldX/Rosetta predict 10 mutations as stabilizing
3-5 will actually be neutral or destabilizing
You waste lab time testing them
Real-World Case Study
Published study: Stabilizing T4 lysozyme
Goal: Find stabilizing mutations using FoldX
FoldX predictions: 20 mutations with ΔΔG < -1 kcal/mol (predicted stabilizing)
Experimental testing: Expressed and measured Tm for all 20
Results:
8 mutations: Stabilizing (+2 to +8°C Tm) ✓
7 mutations: Neutral (±1°C Tm) ✗
5 mutations: Destabilizing (-2 to -5°C Tm) ✗
Success rate: 40%
Cost:
20 mutations × $500 per construct (gene synthesis + expression + purification) = $10,000
12 mutations wasted = $6,000
The problem: FoldX can't distinguish high-confidence from low-confidence predictions.
Why False Positives Happen
Reason 1: Coarse energy function
FoldX uses ~10 energy terms (van der Waals, electrostatics, solvation, etc.)
Real protein energetics: 1000+ atom-atom interactions
Simplifications introduce errors
Reason 2: No structural relaxation
FoldX uses rigid backbone (doesn't allow protein to adjust)
Mutation causes clash → large positive ΔΔG → predicted destabilizing
Reality: Protein backbone shifts slightly, clash resolved → actually neutral
FoldX overestimates destabilization
Reason 3: Missing entropy
FoldX estimates entropy changes, but it's hard
Entropy often dominates small ΔΔG values
Errors in entropy → errors in ΔΔG
Reason 4: Training data bias
FoldX energy function tuned on limited dataset (mostly mesophilic proteins)
Doesn't generalize well to thermophiles, membrane proteins, antibodies
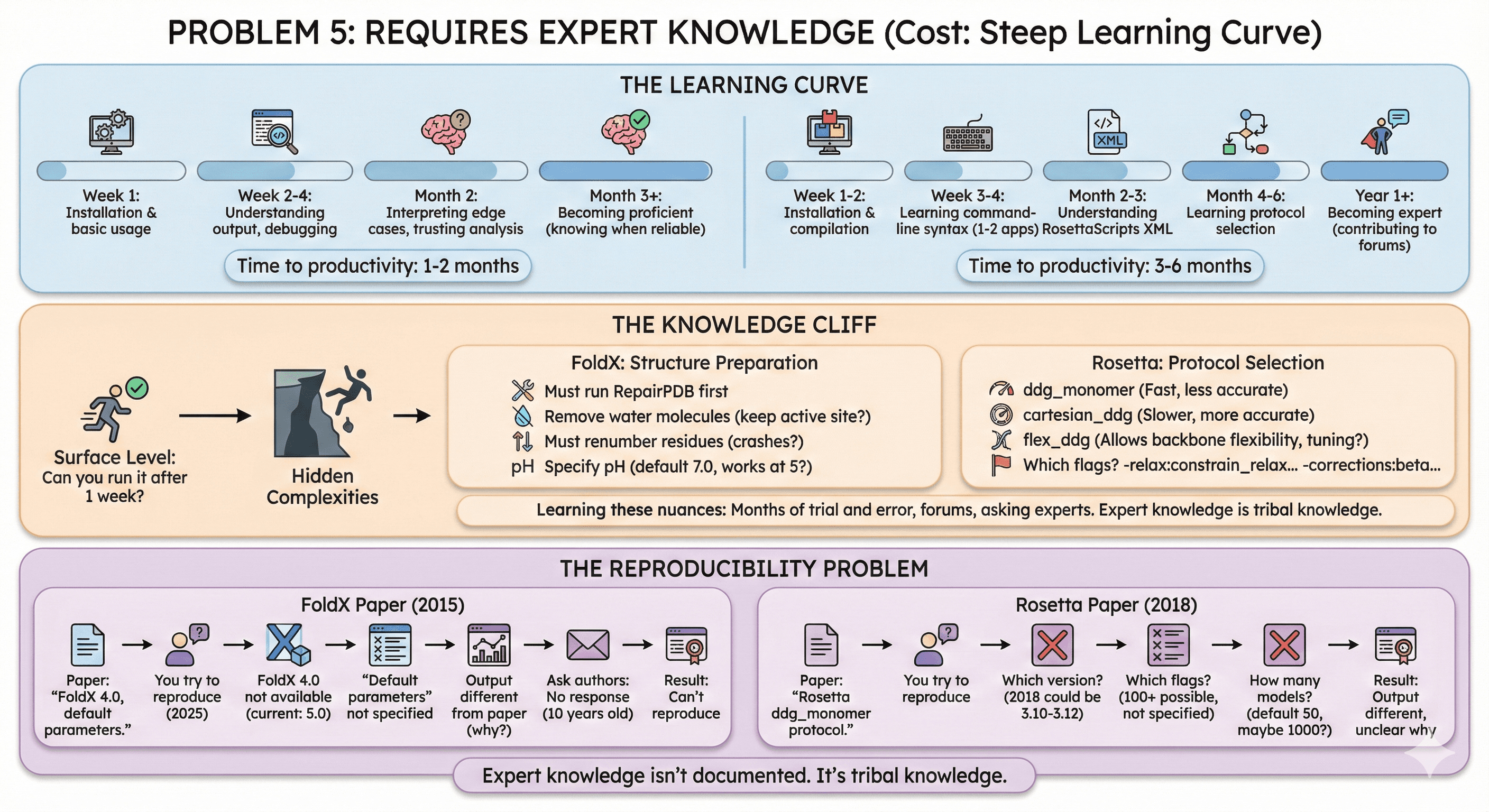
Problem 5: Requires Expert Knowledge (Cost: Steep Learning Curve)
The Learning Curve
FoldX:
Week 1: Installation and basic usage
Week 2-4: Understanding output, debugging common errors
Month 2: Learning which analyses to trust, how to interpret edge cases
Month 3+: Becoming proficient (knowing when predictions are reliable)
Rosetta:
Week 1-2: Installation and compilation
Week 3-4: Learning command-line syntax for 1-2 applications
Month 2-3: Understanding RosettaScripts XML files
Month 4-6: Learning which protocols to use for which tasks
Year 1+: Becoming expert (contributing to Rosetta community forums)
Time to productivity:
FoldX: 1-2 months
Rosetta: 3-6 months
The Knowledge Cliff
You can run FoldX/Rosetta after 1 week. But can you trust the results?
Hidden complexities:
FoldX: Structure preparation
Must run
RepairPDBfirst to fix structureMust remove water molecules (but keep crystallographic waters near active site?)
Must renumber residues (but FoldX sometimes crashes on renumbering)
Must specify pH (default 7.0, but what if your protein works at pH 5?)
Rosetta: Protocol selection
ddg_monomer: Fast, less accuratecartesian_ddg: Slower, more accurate (but when to use?)flex_ddg: Allows backbone flexibility (but how much? requires tuning)Which flags to use?
-relax:constrain_relax_to_start_coords?-corrections:beta_nov16?
Learning these nuances: Months of trial and error, reading forums, asking experts
The Reproducibility Problem
FoldX paper (2015): "ΔΔG calculated using FoldX 4.0 with default parameters."
You try to reproduce (2025):
FoldX 4.0 no longer available (current version: 5.0)
"Default parameters" not specified in paper
Output different from paper (why?)
Ask paper authors: No response (paper 10 years old)
Result: Can't reproduce
Rosetta paper (2018): "Stability calculated using Rosetta ddg_monomer protocol."
You try to reproduce:
Which Rosetta version? (2018 could be 3.10-3.12, different results)
Which flags? (100+ possible flags, paper doesn't specify)
How many models? (default 50, but paper may have used 1000)
Result: Output different, unclear why
Expert knowledge isn't documented. It's tribal knowledge.
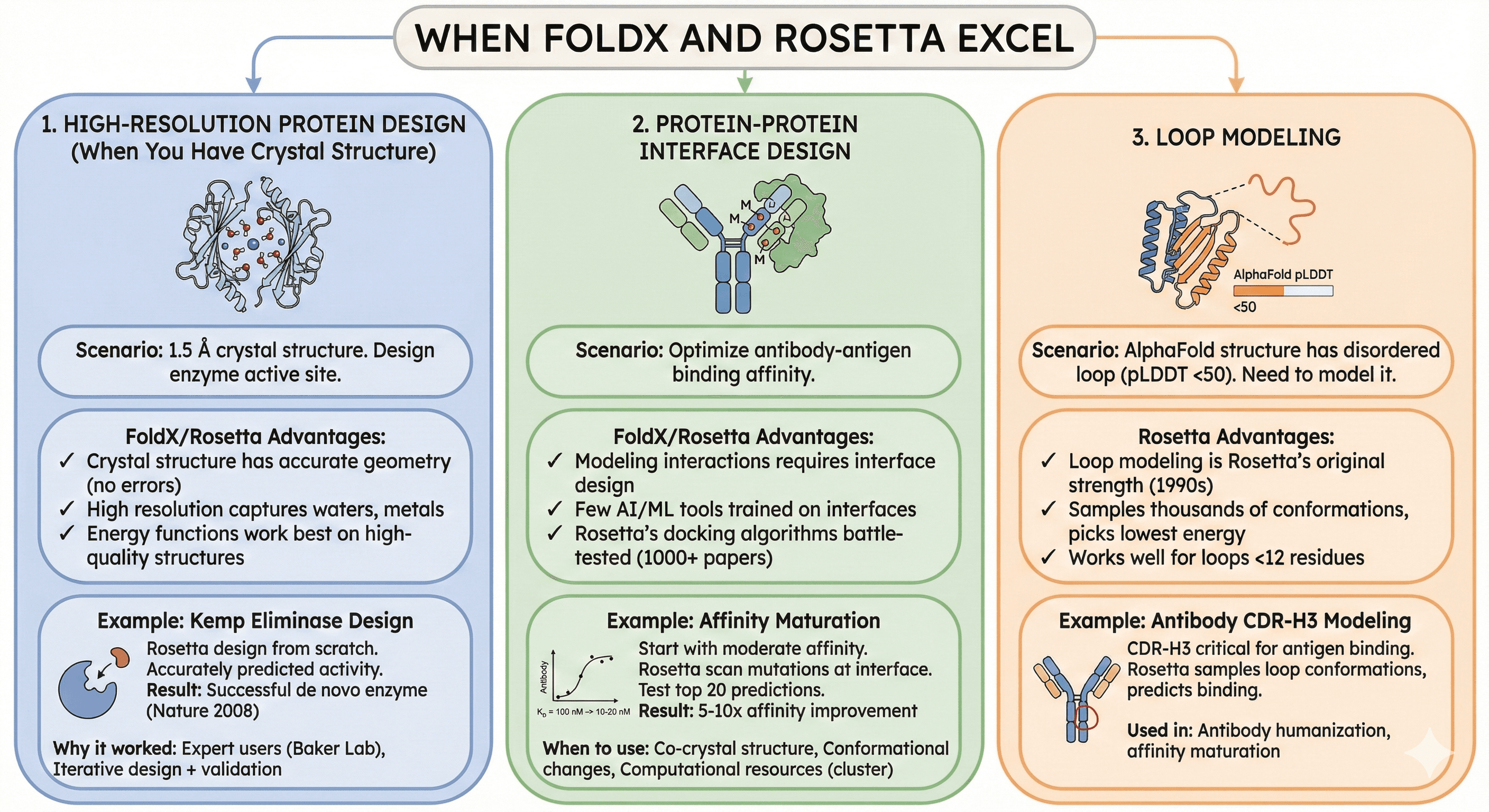
When FoldX and Rosetta Excel
Use Case 1: High-Resolution Protein Design (When You Have Crystal Structure)
Scenario: You have 1.5 Å crystal structure, want to design enzyme active site
FoldX/Rosetta advantages:
Crystal structure has accurate geometry (no model errors)
High resolution captures water molecules, metal ions
Energy functions work best on high-quality structures
Example: Kemp eliminase design
Used Rosetta to design enzyme from scratch
Started with known scaffold, designed active site
Rosetta accurately predicted catalytic activity
Result: Successful de novo enzyme (published Nature 2008)
Why it worked:
Expert users (Baker Lab)
High-quality starting structures
Iterative design + experimental validation
Use Case 2: Protein-Protein Interface Design
Scenario: Optimize antibody-antigen binding affinity
FoldX/Rosetta advantages:
Interface design requires modeling protein-protein interactions
Few AI/ML tools trained on interface data (most focus on monomers)
Rosetta's docking algorithms battle-tested (1000+ papers)
Example: Affinity maturation
Start with moderate-affinity antibody (KD = 100 nM)
Use Rosetta to scan mutations at interface
Test top 20 predictions experimentally
Result: 5-10x affinity improvement
When to use:
You have co-crystal structure of complex
You need to model conformational changes upon binding
You have computational resources (cluster)
Use Case 3: Loop Modeling
Scenario: Your AlphaFold structure has disordered loop (pLDDT <50), you need to model it
Rosetta advantages:
Loop modeling is Rosetta's original strength (1990s)
Samples thousands of conformations, picks lowest energy
Works well for loops <12 residues
Example: Antibody CDR-H3 modeling
CDR-H3 (complementarity-determining region) varies in length/sequence
Critical for antigen binding
Rosetta samples loop conformations, predicts binding
Used in: Antibody humanization, affinity maturation
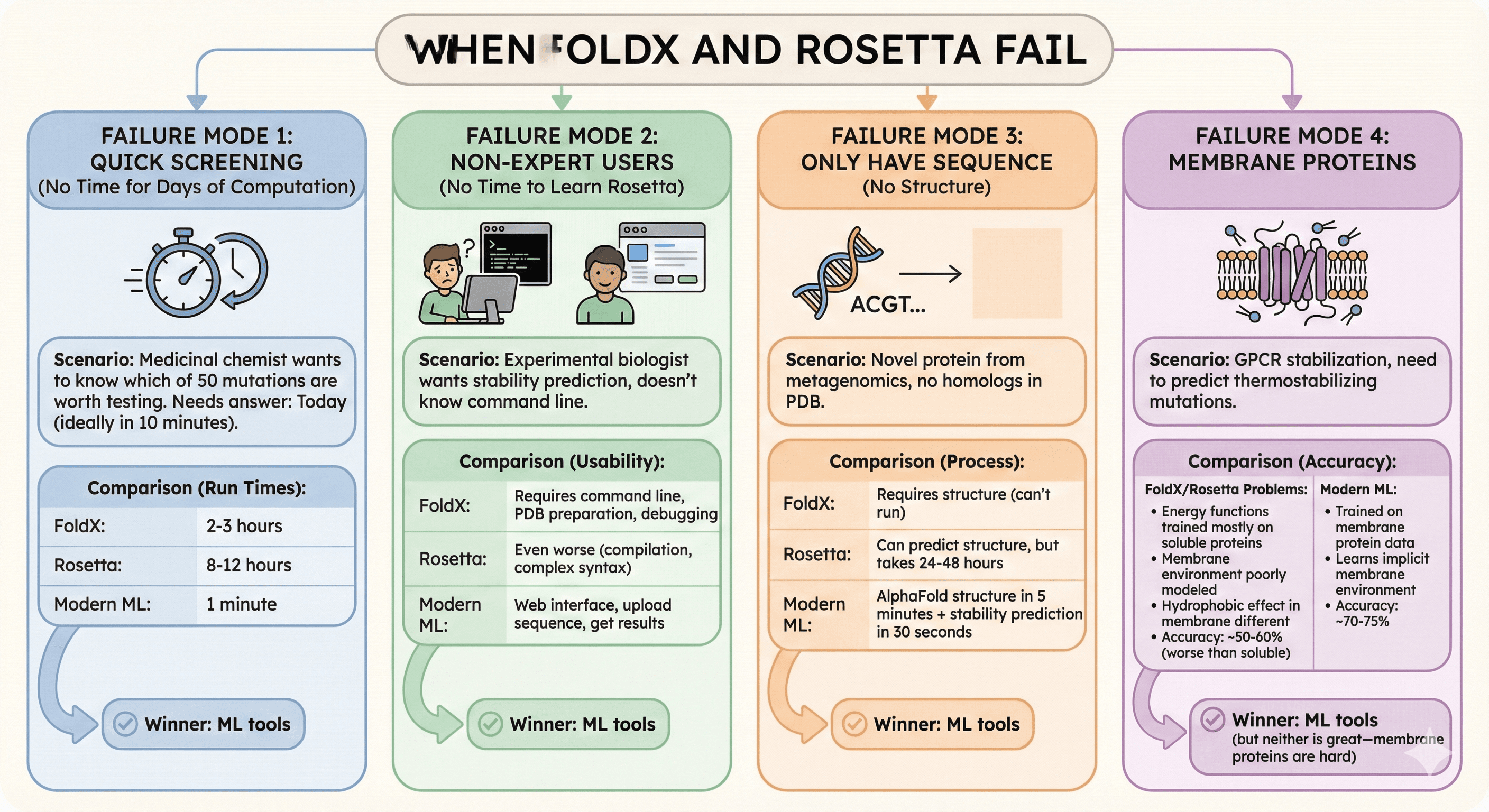
When FoldX and Rosetta Fail
Failure Mode 1: Quick Screening (No Time for Days of Computation)
Scenario: Medicinal chemist wants to know which of 50 mutations are worth testing
Needs answer: Today (ideally in 10 minutes)
FoldX: 2-3 hours
Rosetta: 8-12 hours
Modern ML: 1 minute
Failure Mode 2: Non-Expert Users (No Time to Learn Rosetta)
Scenario: Experimental biologist wants stability prediction, doesn't know command line
FoldX: Requires command line, PDB preparation, debugging
Rosetta: Even worse (compilation, complex syntax)
Modern ML: Web interface, upload sequence, get results
Failure Mode 3: Only Have Sequence (No Structure)
Scenario: Novel protein from metagenomics, no homologs in PDB
FoldX: Requires structure (can't run)
Rosetta: Can predict structure, but takes 24-48 hours
Modern ML: AlphaFold structure in 5 minutes + stability prediction in 30 seconds
Failure Mode 4: Membrane Proteins
Scenario: GPCR stabilization, need to predict thermostabilizing mutations
FoldX/Rosetta problems:
Energy functions trained mostly on soluble proteins
Membrane environment poorly modeled (lipid bilayer, detergents)
Hydrophobic effect in membrane different from solution
Accuracy: ~50-60% (worse than soluble proteins)
Modern ML:
Trained on membrane protein data (AlphaFold saw membrane proteins)
Learns implicit membrane environment
Accuracy: ~70-75%
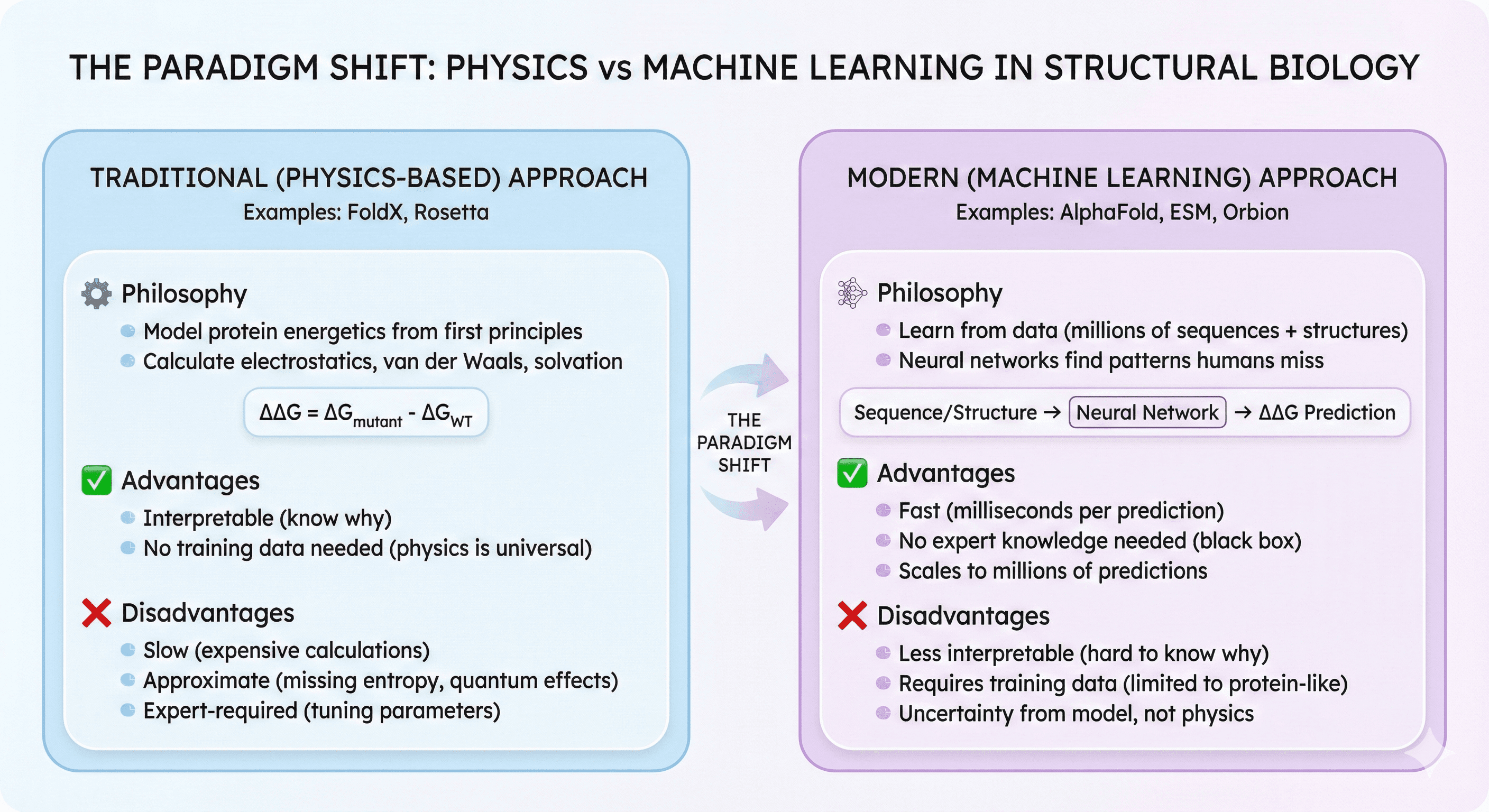
The Paradigm Shift: Physics vs Machine Learning
Traditional (Physics-Based) Approach
FoldX/Rosetta philosophy:
Model protein energetics from first principles
Calculate electrostatics, van der Waals, solvation
ΔΔG = ΔG_mutant - ΔG_WT
Advantages:
Interpretable (know why mutation is stabilizing)
No training data needed (physics is universal)
Disadvantages:
Slow (expensive calculations)
Approximate (missing entropy, quantum effects)
Expert-required (tuning parameters)
Modern (Machine Learning) Approach
AlphaFold/ESM/Orbion philosophy:
Learn from data (millions of protein sequences + structures)
Neural networks find patterns humans miss
Predict ΔΔG directly from sequence/structure
Advantages:
Fast (milliseconds per prediction)
No expert knowledge needed (black box)
Scales to millions of predictions
Disadvantages:
Less interpretable (hard to know why)
Requires training data (limited to protein-like sequences)
Uncertainty from model, not physics
The Accuracy Comparison (Literature Benchmarks)
Method | Correlation with experiment (R) | Accuracy (% correct direction) | Speed (per mutation) | Requires structure? |
|---|---|---|---|---|
FoldX | 0.6-0.7 | 65-70% | 2-5 min | Yes (PDB) |
Rosetta ddg_monomer | 0.5-0.7 | 60-70% | 10-30 min | Yes (PDB) |
Rosetta cartesian_ddg | 0.6-0.75 | 70-75% | 30-60 min | Yes (PDB) |
ESM-1v (2021) | 0.7-0.8 | 75-80% | <1 sec | No (sequence) |
AlphaFold2 + ΔΔG (2022) | 0.65-0.75 | 70-75% | 5-30 sec | No (sequence) |
Orbion AstraSTASIS | 0.75-0.85 | 75-85% | <1 sec | No (sequence) |
Key insight: Modern ML tools are faster AND more accurate than traditional tools.
Key Takeaway
FoldX and Rosetta were revolutionary 20 years ago. They're still powerful for specialized tasks (high-resolution design, interface optimization, loop modeling). But for most users, they've become bottlenecks:
The 5 problems:
Installation hell: Days of setup time (dependency issues, compilation)
Slow computation: Hours to days per analysis (doesn't scale)
No uncertainty: Single ΔΔG value (no confidence interval) → false confidence
High false positives: 30-50% of "stabilizing" predictions fail experimentally
Expert-required: Months to learn, tribal knowledge needed
When to use traditional tools:
You're an expert (know the pitfalls)
You have crystal structure (high resolution)
You need interpretability (why mutation works)
You're doing interface design or loop modeling
When to use modern ML:
You want fast results (seconds not hours)
You're non-expert (no time to learn Rosetta)
You only have sequence (no structure)
You need confidence scores (prioritize experiments)

