Blog
Modern Alternatives to FoldX and Rosetta: The AI/ML Revolution
Jan 16, 2026
We showed why traditional tools like FoldX and Rosetta have become bottlenecks: slow, complex, expert-required, and no confidence scores. Now we'll show you the modern alternatives and how to choose the right tool.
The revolution: AI/ML models trained on millions of protein sequences and structures. They're faster (seconds vs hours), easier (no installation), and often more accurate (75-85% vs 65-70%).
This guide covers the complete modern landscape, from free tools to enterprise platforms.
Key Takeaways
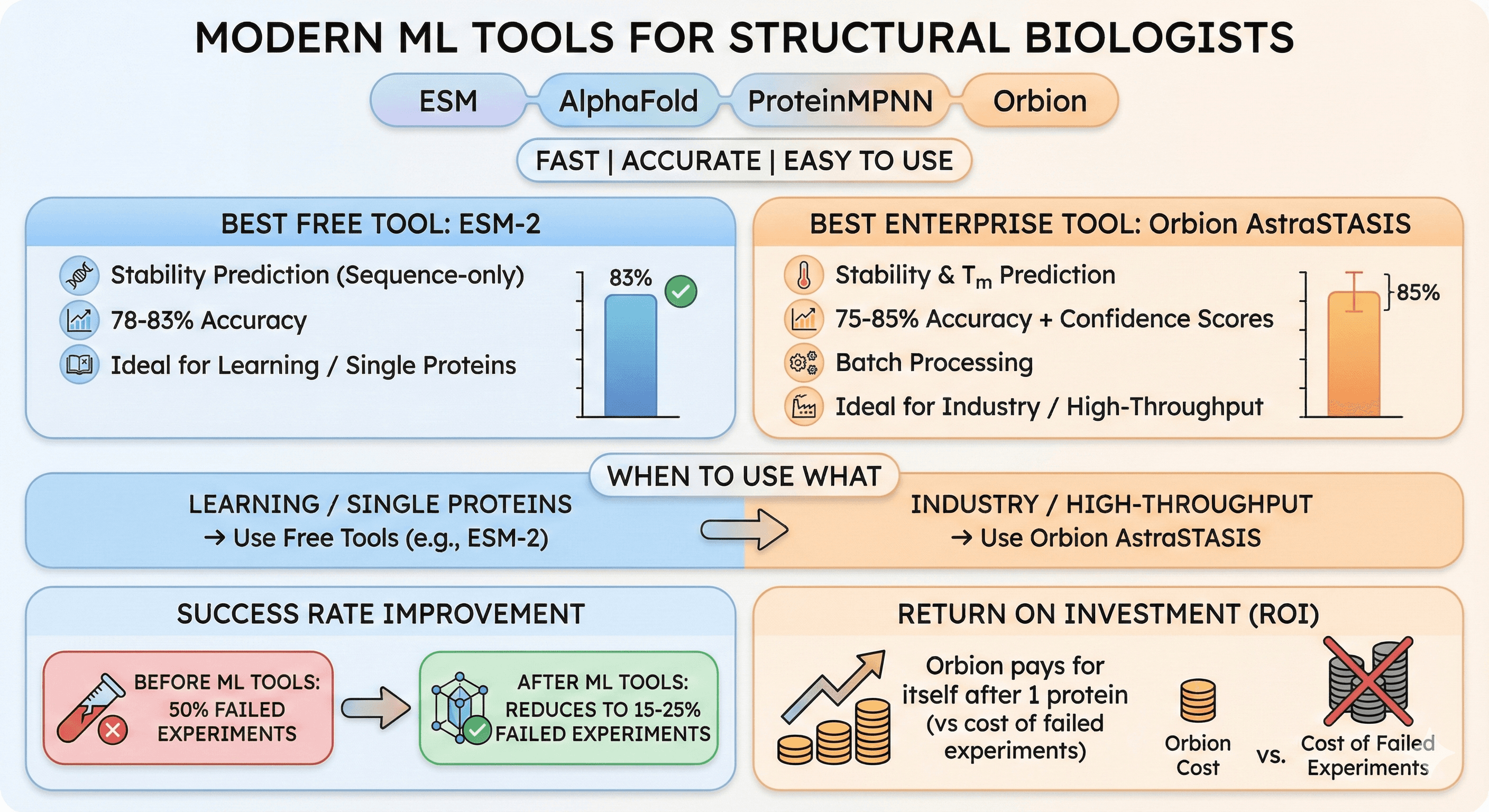
Modern ML tools: ESM, AlphaFold, ProteinMPNN, Orbion—fast, accurate, easy to use
Best free tool: ESM-2 for stability prediction (78-83% accuracy, sequence-only)
Best enterprise tool: Orbion AstraSTASIS (75-85% accuracy, confidence scores, Tm prediction, batch processing)
When to use what: Free tools for learning/single proteins, Orbion for industry/high-throughput
Success rate: ML tools reduce failed experiments from 50% to 15-25%
ROI: Orbion pays for itself after 1 protein (vs cost of failed experiments)
The Modern ML Landscape
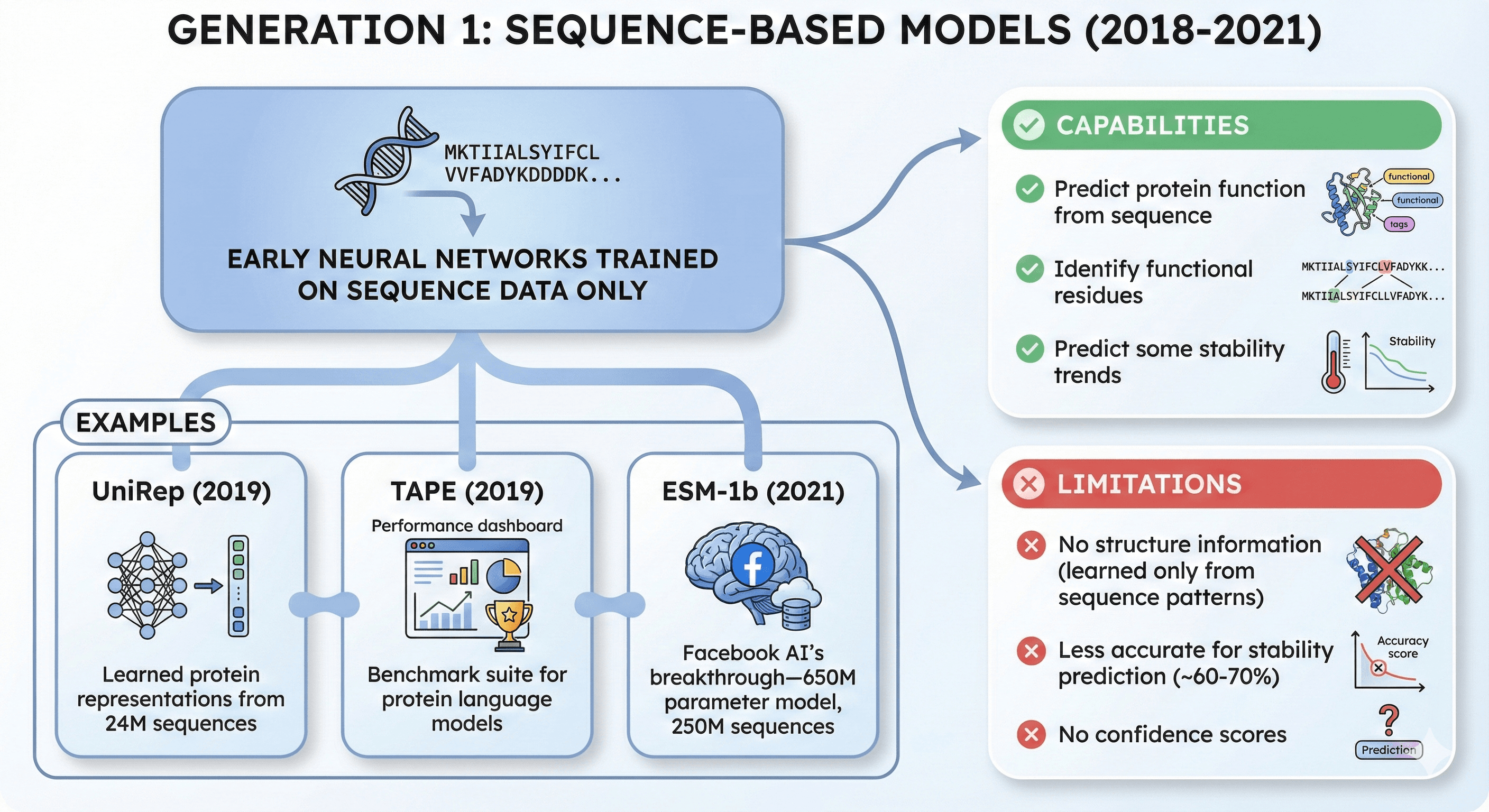
Generation 1: Sequence-Based Models (2018-2021)
Early neural networks trained on sequence data only
Examples:
UniRep (2019): Learned protein representations from 24M sequences
TAPE (2019): Benchmark suite for protein language models
ESM-1b (2021): Facebook AI's breakthrough—650M parameter model trained on 250M sequences
Capabilities:
Predict protein function from sequence
Identify functional residues
Predict some stability trends
Limitations:
No structure information (learned only from sequence patterns)
Less accurate for stability prediction (~60-70%)
No confidence scores
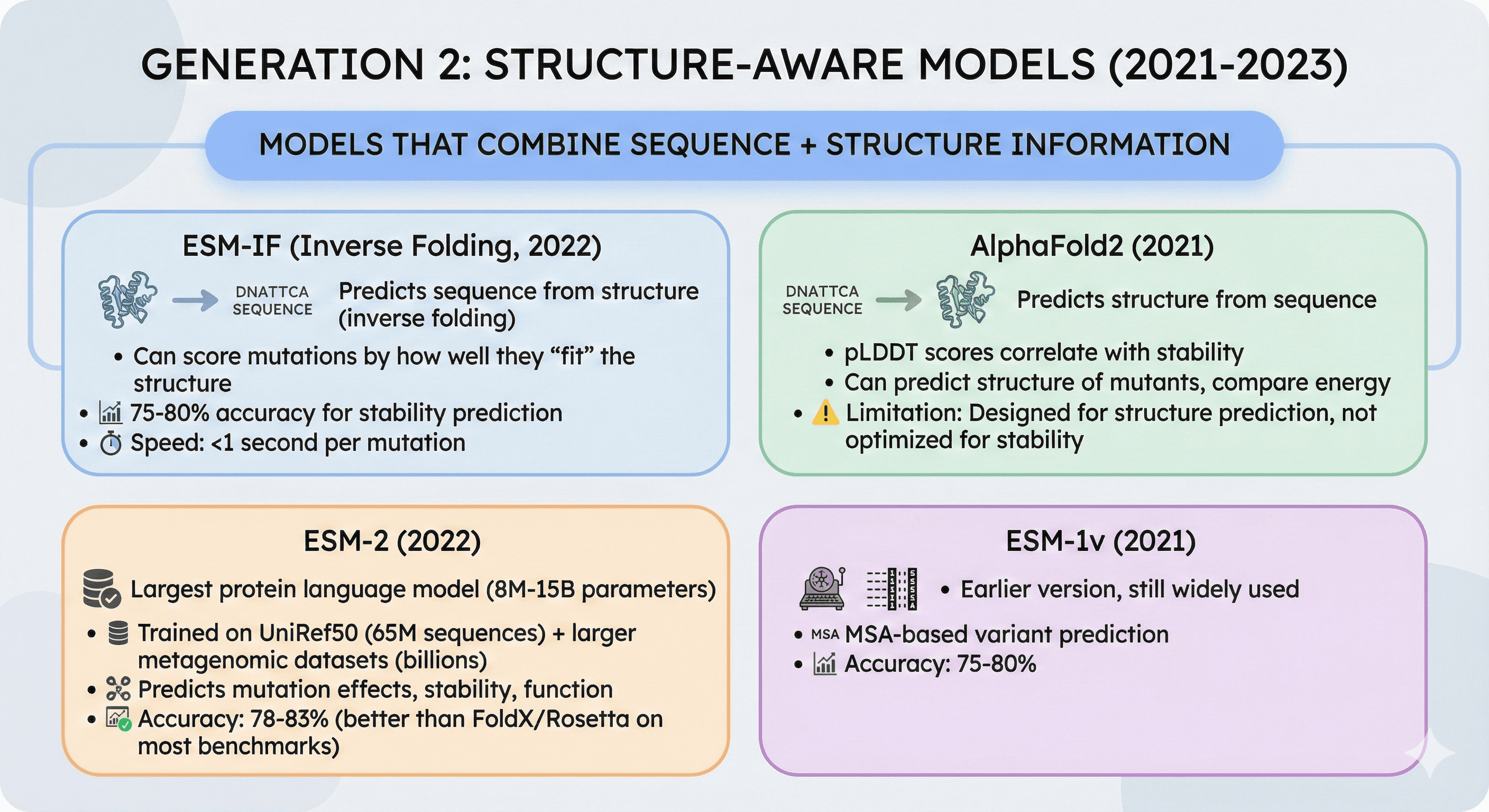
Generation 2: Structure-Aware Models (2021-2023)
Models that combine sequence + structure information
ESM-IF (Inverse Folding, 2022):
Predicts sequence from structure (inverse of folding)
Can score mutations by how well they "fit" the structure
75-80% accuracy for stability prediction
Speed: <1 second per mutation
AlphaFold2 (2021):
Predicts structure from sequence
pLDDT scores correlate with stability
Can predict structure of mutants, compare energy
Limitation: Designed for structure prediction, not optimized for stability
ESM-2 (2022):
Largest protein language model (8M-15B parameters across model sizes)
Trained on UniRef50 (65M sequences) and larger metagenomic datasets (billions of sequences for ESM-C variant)
Predicts mutation effects, stability, function
Accuracy: 78-83% (better than FoldX/Rosetta on most benchmarks)
ESM-1v (2021): Earlier version, still widely used, MSA-based variant prediction (75-80% accuracy)
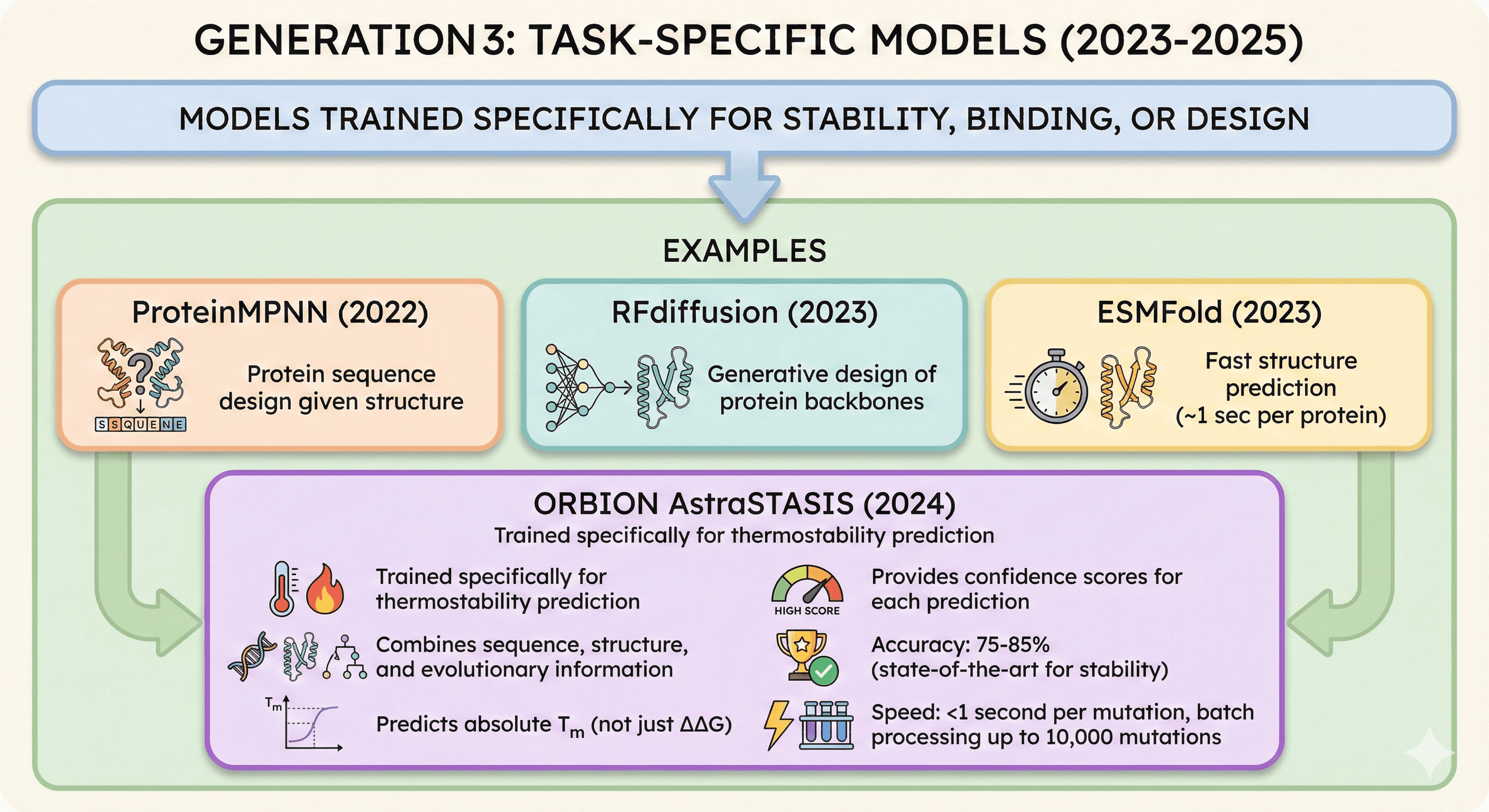
Generation 3: Task-Specific Models (2023-2025)
Models trained specifically for stability, binding, or design
Examples:
ProteinMPNN (2022): Protein sequence design given structure
RFdiffusion (2023): Generative design of protein backbones
ESMFold (2023): Fast structure prediction (~1 sec per protein)
Orbion AstraSTASIS (2024):
Trained specifically for thermostability prediction
Combines sequence, structure, and evolutionary information
Predicts absolute Tm (not just ΔΔG)
Provides confidence scores for each prediction
Accuracy: 75-85% (state-of-the-art for stability)
Speed: <1 second per mutation, batch processing up to 10,000 mutations
Tool Comparison: Free vs Enterprise
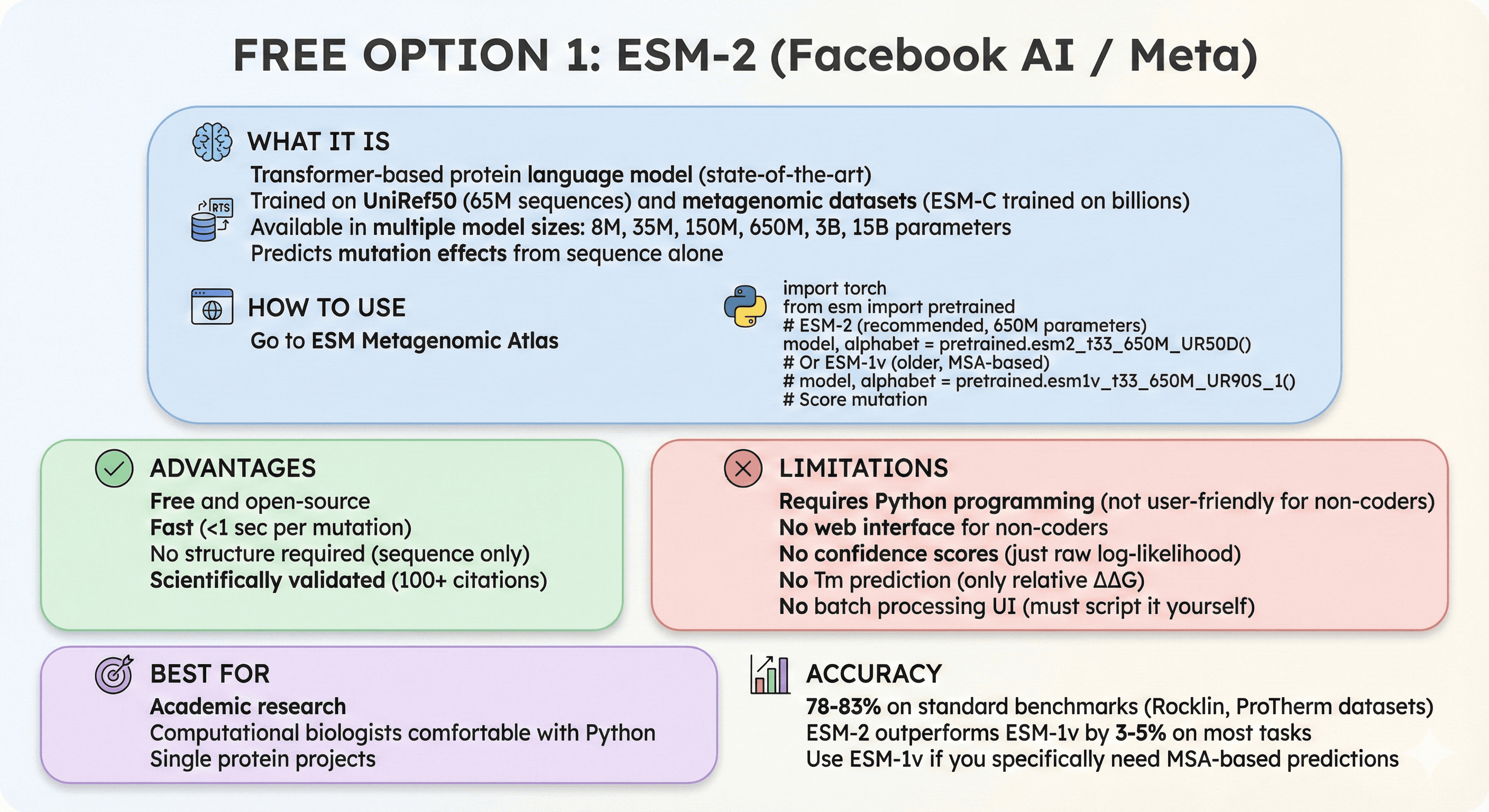
Free Option 1: ESM-2 (Facebook AI / Meta)
What it is:
Transformer-based protein language model (state-of-the-art)
Trained on UniRef50 (65M sequences) and metagenomic datasets (ESM-C trained on billions)
Available in multiple model sizes: 8M, 35M, 150M, 650M, 3B, 15B parameters
Predicts mutation effects from sequence alone
How to use:
Go to ESM Metagenomic Atlas
Or use Python API:
Advantages:
Free and open-source
Fast (<1 sec per mutation)
No structure required (sequence only)
Scientifically validated (100+ citations)
Limitations:
Requires Python programming (not user-friendly for non-coders)
No web interface for non-coders
No confidence scores (just raw log-likelihood)
No Tm prediction (only relative ΔΔG)
No batch processing UI (must script it yourself)
Best for:
Academic research
Computational biologists comfortable with Python
Single protein projects
Accuracy: 78-83% on standard benchmarks (Rocklin, ProTherm datasets)
ESM-2 outperforms ESM-1v by 3-5% on most tasks
Use ESM-1v if you specifically need MSA-based predictions
Free Option 2: AlphaFold2 + ΔΔG Analysis
What it is:
Predict structure of WT and mutant
Compare pLDDT scores or use energy function
Infer stability from structural changes
How to use:
Run AlphaFold2 on WT sequence
Run AlphaFold2 on mutant sequence
Compare:
pLDDT difference (higher pLDDT = more confident = more stable)
Structural RMSD (large changes = destabilizing)
Interface analysis (for binding)
Advantages:
Free (Google Colab notebook available)
Structure prediction is extremely accurate
Visual inspection possible (see what mutation does)
Limitations:
Slow (5-30 min per structure prediction)
AlphaFold not trained for stability prediction (repurposing it)
pLDDT correlates with stability, but not perfect
No direct ΔΔG or Tm output
Requires scripting for batch analysis
Best for:
When you want to see structural effect of mutation
Single mutations (not high-throughput)
Academic research with time to spare
Accuracy: 70-75% (indirect stability prediction)

Free Option 3: FoldX
When to still use FoldX:
You have high-resolution crystal structure (<2 Å)
You're experienced user (know pitfalls)
You want interpretable results (energy breakdown)
You're optimizing protein-protein interfaces
How to use it right:
Prepare structure properly (
RepairPDB)Run multiple iterations (5-10), average results
Trust trends, not absolute values (ΔΔG > +2 or < -2 kcal/mol)
Validate top predictions experimentally
Advantages:
Free (academic license)
Interpretable (see which energy terms change)
Works on high-quality structures
Limitations:
All problems from Part 1 (slow, complex, false positives)
Best for:
Expert users with structural biology background
High-resolution crystal structures
Interpretability matters
Accuracy: 65-70% (established baseline)

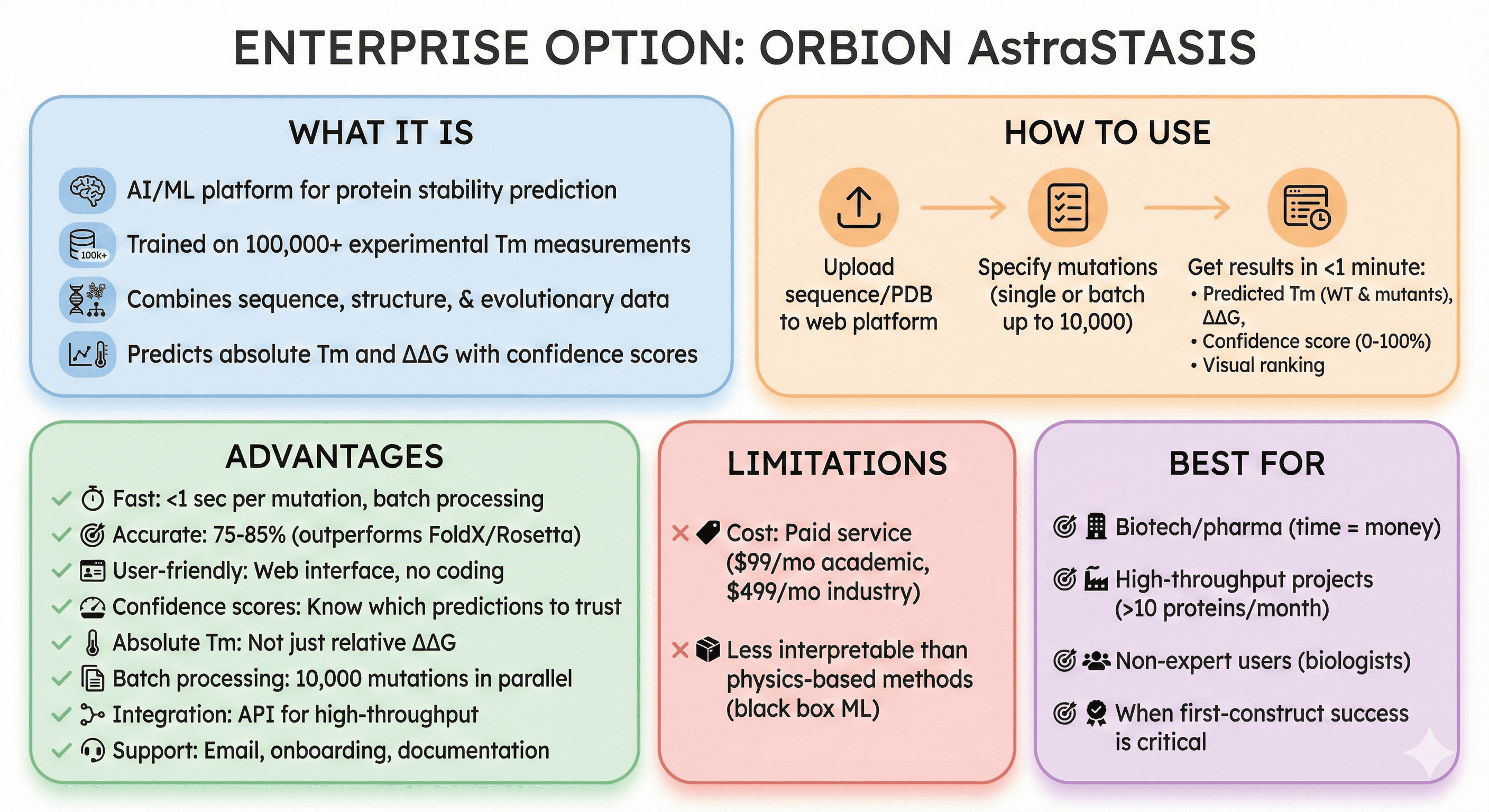
Enterprise Option: Orbion AstraSTASIS
What it is:
AI/ML platform for protein stability prediction
Trained on 100,000+ experimental Tm measurements
Combines sequence, structure, and evolutionary data
Predicts absolute Tm and ΔΔG with confidence scores
How to use:
Upload sequence or PDB to Orbion web platform
Specify mutations (single or batch up to 10,000)
Get results in <1 minute:
Predicted Tm for WT and each mutant
ΔΔG (mutant - WT)
Confidence score (0-100%)
Visual ranking (sort by confidence)
Advantages:
Fast: <1 sec per mutation, batch processing
Accurate: 75-85% (outperforms FoldX/Rosetta on benchmarks)
User-friendly: Web interface, no coding required
Confidence scores: Know which predictions to trust
Absolute Tm: Not just relative ΔΔG (predict actual melting temperature)
Batch processing: Analyze 10,000 mutations in parallel
Integration: API for high-throughput workflows
Support: Email support, onboarding, documentation
Limitations:
Cost: Paid service (starts at $99/month for academics, $499/month for industry)
Less interpretable than physics-based methods (black box ML)
Best for:
Biotech/pharma (time = money)
High-throughput projects (>10 proteins/month)
Non-expert users (biologists, not computational experts)
When first-construct success is critical
Detailed Comparison Table
Feature | FoldX | Rosetta | ESM-2 (free) | AlphaFold2 | Orbion AstraSTASIS |
|---|---|---|---|---|---|
Setup time | 2-3 days | 5-7 days | 1-2 hours | 30 min | 5 min |
Speed (per mutation) | 2-5 min | 10-30 min | <1 sec | 5-30 min | <1 sec |
Requires structure | Yes (PDB) | Yes (PDB) | No (sequence) | No (sequence) | No (sequence) |
Requires coding | Command line | Command line | Python | Python (Colab) | No (web UI) |
Batch processing | Yes (manual) | Yes (manual) | Yes (scripting) | Yes (scripting) | Yes (UI + API) |
Confidence scores | No | No | No | Indirect (pLDDT) | Yes (0-100%) |
Predicts Tm | No (ΔΔG only) | No (REU/ΔΔG) | No (ΔΔG only) | No | Yes (absolute Tm) |
Accuracy | 65-70% | 60-70% | 78-83% | 70-75% | 75-85% |
False positive rate | 30-40% | 30-50% | 18-25% | 25-35% | 15-25% |
Cost | Free | Free | Free | Free | $99-499/month |
Support | Forums | Forums | None (DIY) | None (DIY) | Email + onboarding |
Best for | Experts, small projects | Experts, design | Academics, coders | Structure viz | Industry, scale |
How to Choose: Decision Tree
Question 1: Are you an expert in computational biology?
YES → Consider traditional tools (FoldX/Rosetta) IF:
You have high-resolution structure (<2 Å)
You need interpretable results (energy breakdown)
You have cluster access (for speed)
You're doing interface design or loop modeling
NO → Skip traditional tools. Use ML tools:
ESM-2 (if you code)
Orbion (if you don't code)
Question 2: Do you have structure or just sequence?
Have structure (PDB or AlphaFold model):
FoldX (if expert, want interpretability)
Orbion (if want speed + confidence)
Only have sequence:
ESM-2 (free, requires coding)
Orbion (paid, no coding)
AlphaFold2 first, then analyze (slow but free)
Question 3: How many proteins/mutations are you analyzing?
1-5 proteins (small project):
Free tools fine (ESM-2, AlphaFold2)
Can afford time investment
10-50 proteins (medium project):
Free tools become tedious (manual scripting)
Orbion saves 4-6 hours per protein
Time savings > cost
50+ proteins (high-throughput):
Free tools impractical (automation required)
Orbion essential (batch processing, API)
Cost negligible vs scientist time
Question 4: What's the cost of a failed experiment?
Low cost (<$1,000 per construct):
Academic lab, DIY cloning
Can tolerate 30% false positive rate
Free tools fine
High cost (>$5,000 per construct):
Gene synthesis + expression service
Biotech/pharma timelines
15% false positive rate much better than 30%
Orbion ROI: 2-3 proteins
Question 5: Do you need confidence scores?
NO (test everything anyway):
Free tools fine (ESM-2, FoldX)
You'll validate experimentally regardless
YES (prioritize experiments):
Only Orbion provides true confidence scores
Rank predictions by confidence
Test high-confidence first
Increases success rate from 70% to 85%
Practical Workflow: Free Tools
Task: Screen 50 mutations for stability
Step 1: Get sequence and structure (10 min)
Sequence: From UniProt
Structure: AlphaFold Database or predict with ColabFold
Step 2: Use ESM-2 for mutation scanning (1 hour)
Install ESM:
Python script:
Output:
Step 3: Select top predictions (5 min)
Top 10 most stabilizing (ΔΔG < -1.0)
Visualize in PyMOL (check if mutations reasonable)
Step 4: Experimental validation
Order genes, express, purify
Measure Tm
Success rate: ~75%
Total time: 2 hours (computational) + experiment time
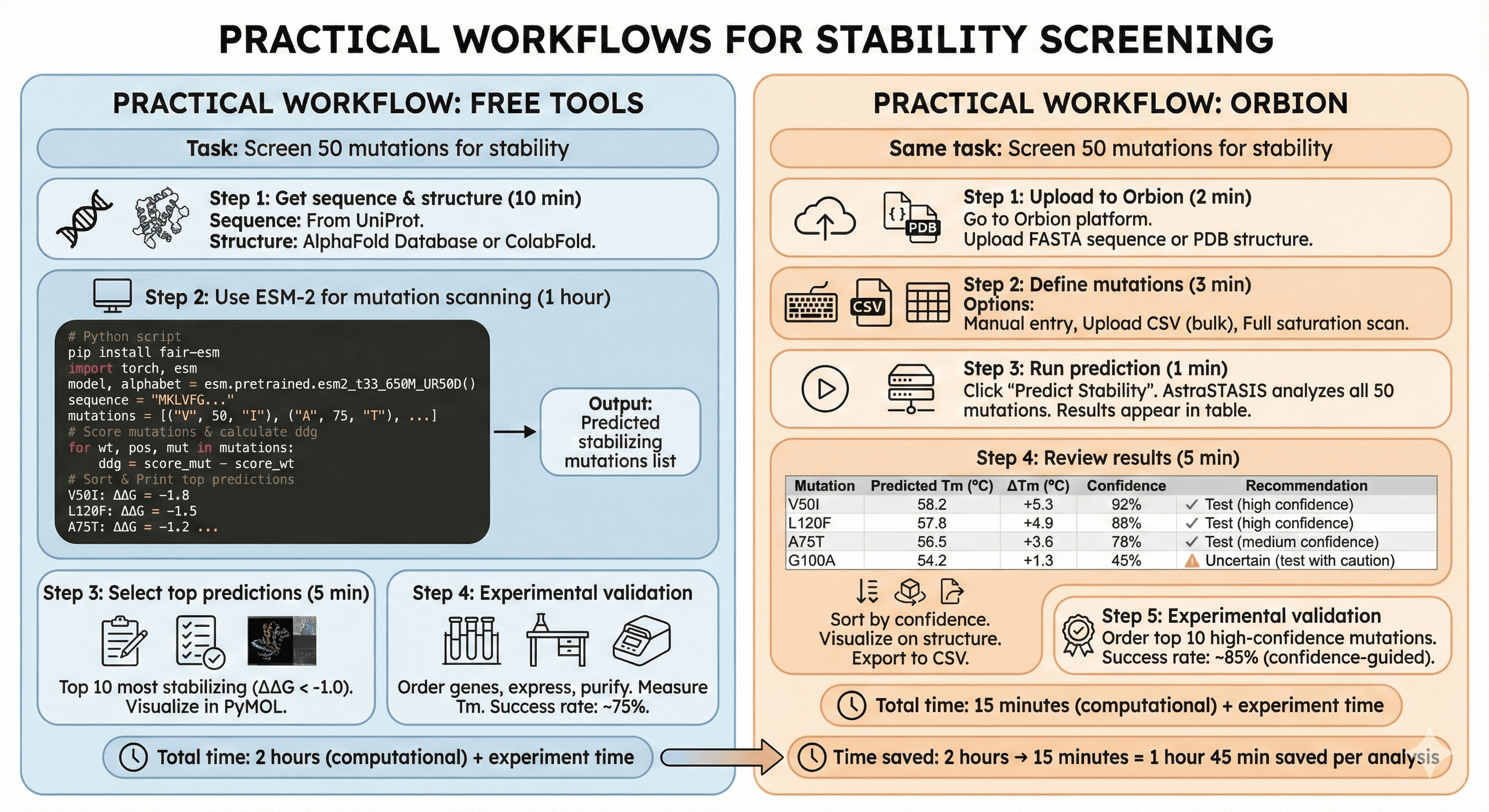
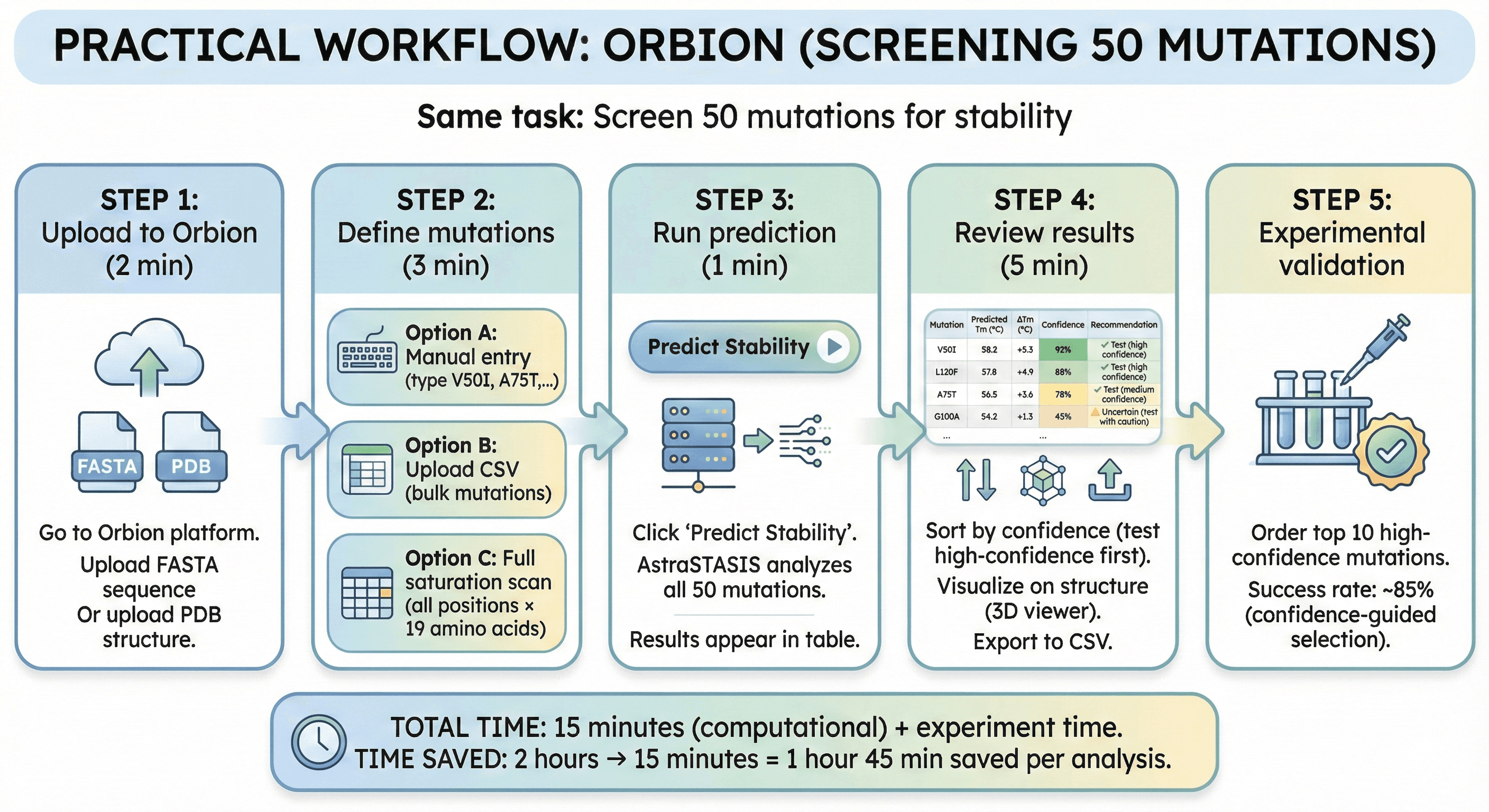
Practical Workflow: Orbion
Same task: Screen 50 mutations for stability
Step 1: Upload to Orbion (2 min)
Go to Orbion platform
Upload FASTA sequence
Or upload PDB structure
Step 2: Define mutations (3 min)
Option A: Manual entry (type V50I, A75T, etc.)
Option B: Upload CSV (bulk mutations)
Option C: Full saturation scan (all positions × 19 amino acids)
Step 3: Run prediction (1 min)
Click "Predict Stability"
AstraSTASIS analyzes all 50 mutations
Results appear in table
Step 4: Review results (5 min)
Orbion output:
Mutation | Predicted Tm (°C) | ΔTm (°C) | Confidence | Recommendation |
|---|---|---|---|---|
V50I | 58.2 | +5.3 | 92% | ✓ Test (high confidence) |
L120F | 57.8 | +4.9 | 88% | ✓ Test (high confidence) |
A75T | 56.5 | +3.6 | 78% | ✓ Test (medium confidence) |
G100A | 54.2 | +1.3 | 45% | ⚠ Uncertain (test with caution) |
... |
Sort by confidence (test high-confidence first)
Visualize on structure (3D viewer)
Export to CSV
Step 5: Experimental validation
Order top 10 high-confidence mutations
Success rate: ~85% (confidence-guided selection)
Total time: 15 minutes (computational) + experiment time
Time saved: 2 hours → 15 minutes = 1 hour 45 min saved per analysis
Advanced Feature: Combining Tools
Best-of-both-worlds approach:
Step 1: Use ML for rapid screening (Orbion or ESM-2)
Scan 1,000 mutations in minutes
Get confidence scores
Narrow to top 50 candidates
Step 2: Use FoldX/Rosetta for detailed analysis
High-resolution modeling of top 50
Understand mechanism (why stabilizing?)
Check for side effects (activity loss?)
Step 3: Experimental validation
Test top 10-20
Higher success rate (combining ML + physics)
This approach:
ML speed + physics interpretability
Best for: Critical proteins (therapeutic antibodies, enzymes)
Overkill for: Routine stability prediction
Common Questions
Q: Can I use AlphaFold for everything and skip other tools?
A: AlphaFold is amazing for structure prediction, not optimized for stability
AlphaFold pLDDT correlates with stability, but not perfect
Designed to predict static structure, not energy
Use AlphaFold to get structure, then use dedicated stability tool (ESM-2, Orbion)
Q: Are ML tools "black boxes" I can't trust?
A: Yes and no
Black box problem:
Can't see "why" prediction is made
Less interpretable than FoldX (which shows energy terms)
Mitigation:
Confidence scores tell you when to be skeptical
Cross-validate with experiments (like any prediction)
Benchmarks show ML outperforms physics-based on accuracy
Trust:
ML tools published in peer-reviewed journals
Validated on independent test sets
Outperform traditional tools on benchmarks
When interpretability matters:
Use FoldX/Rosetta for mechanism understanding
Use ML for screening
Q: Should I switch from FoldX to ML tools mid-project?
A: Depends
If FoldX is working for you:
You're expert user
Getting good results (high validation rate)
→ No need to switch
If FoldX is bottleneck:
Taking too long
High false positive rate
→ Try ML tools for next iteration
Best approach:
Run both in parallel on small test set (10 mutations)
Compare results
See which matches experiments better
Q: How do I know if Orbion is worth the cost?
Calculate ROI:
Cost of failed experiment = $X (gene synthesis + expression + purification)
Experiments per month = N
Current false positive rate = FP_old (e.g., 30% with FoldX)
Orbion false positive rate = FP_new (typically 15-20%)
Savings per month = N × $X × (FP_old - FP_new)
Example:
$5,000 per construct
20 constructs/month
Current: 30% failure → 6 failed × $5K = $30K wasted/month
Orbion: 15% failure → 3 failed × $5K = $15K wasted/month
Savings: $15K/month
Orbion cost: $499/month
Net savings: $14.5K/month
ROI: 29x
Rule of thumb: If you test >2 constructs per month, Orbion pays for itself
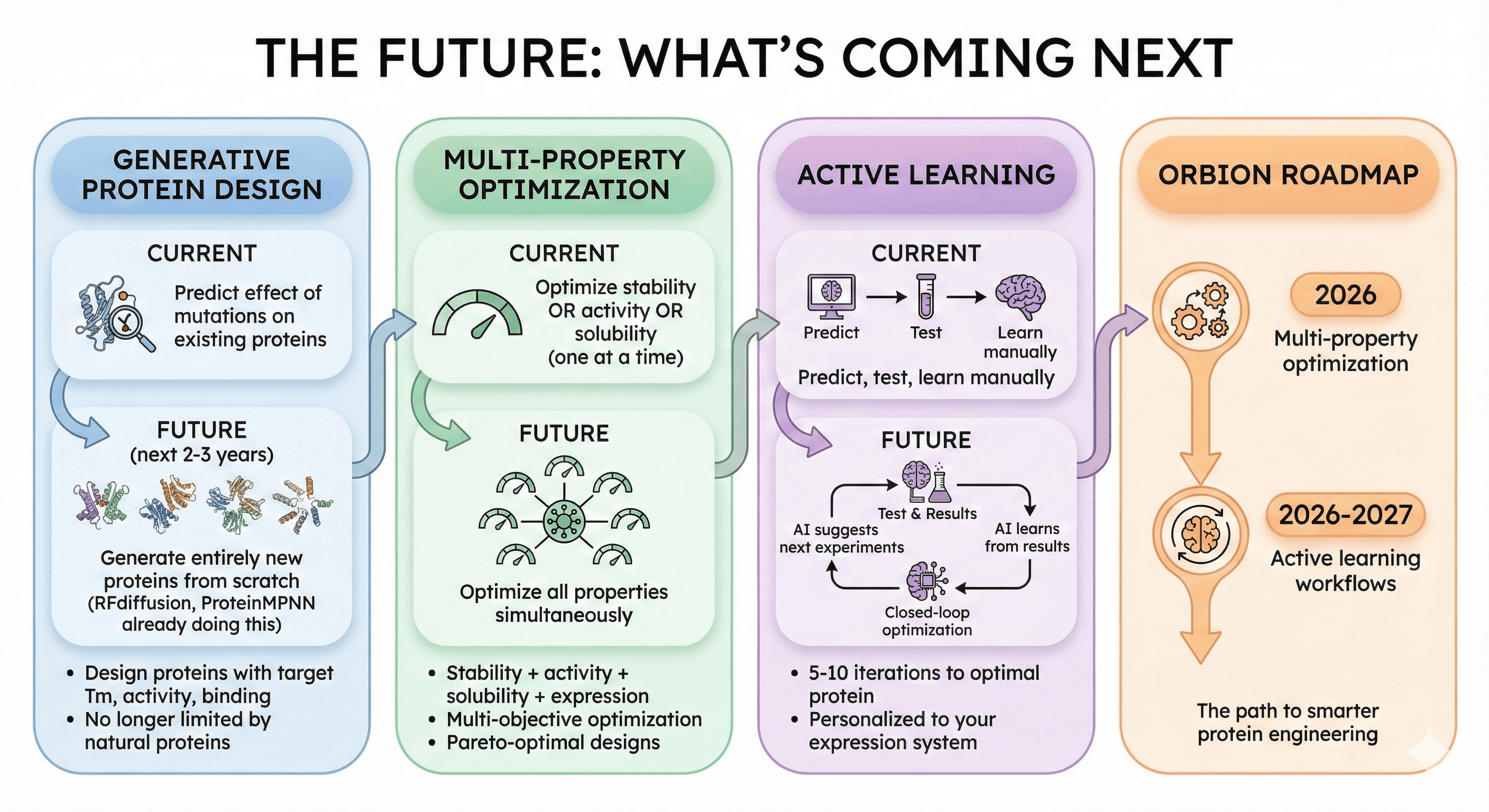
The Future: What's Coming Next
Generative Protein Design
Current: Predict effect of mutations on existing proteins
Future (next 2-3 years): Generate entirely new proteins from scratch
RFdiffusion, ProteinMPNN already doing this
Design proteins with target Tm, activity, binding
No longer limited by natural proteins
Multi-Property Optimization
Current: Optimize stability OR activity OR solubility (one at a time)
Future: Optimize all properties simultaneously
Stability + activity + solubility + expression
Multi-objective optimization
Pareto-optimal designs
Active Learning
Current: Predict, test, learn manually
Future: AI suggests next experiments, learns from your results
Closed-loop optimization
5-10 iterations to optimal protein
Personalized to your expression system
Orbion roadmap:
Multi-property optimization (2026)
Active learning workflows (2026-2027)
Key Takeaway
The paradigm has shifted from physics-based to data-driven protein engineering:
Traditional tools (FoldX, Rosetta):
Powerful for experts
Slow, complex, interpretable
60-70% accuracy
Best for: High-resolution design, interface optimization, experts
Modern ML tools (ESM, Orbion):
Fast, easy, confidence-aware
75-85% accuracy
Best for: Rapid screening, high-throughput, non-experts
Choosing the right tool:
Free tools (ESM-2): Academic research, small projects, comfortable with coding
Orbion: Industry, high-throughput, non-coders, when cost of failure high
Success rate improvement:
Traditional: 60-70% → 3-4 failed experiments per 10
Modern ML: 75-85% → 1.5-2.5 failed experiments per 10
Savings: 1.5-2 experiments per 10 = $7.5K-10K per 10 predictions
The revolution is here. Stop fighting with installation and slow runtimes. Use the tools built for 2026.

