Blog
Why Your Protein Doesn't Express in E. coli (When It Should)
Jan 19, 2026
You cloned the gene. Sequence-verified the construct. Transformed competent cells. Induced with IPTG. And got... nothing. No band on SDS-PAGE. Or worse—a beautiful band in the insoluble fraction, mocking you from the pellet. You've just joined the club that every protein biochemist eventually joins: the E. coli expression failure club.
Here's the thing: E. coli is supposed to be the workhorse. Fast, cheap, scalable. But for 40-60% of eukaryotic proteins, it simply doesn't work (Structural Genomics Consortium data). The question is: why yours?
Key Takeaways
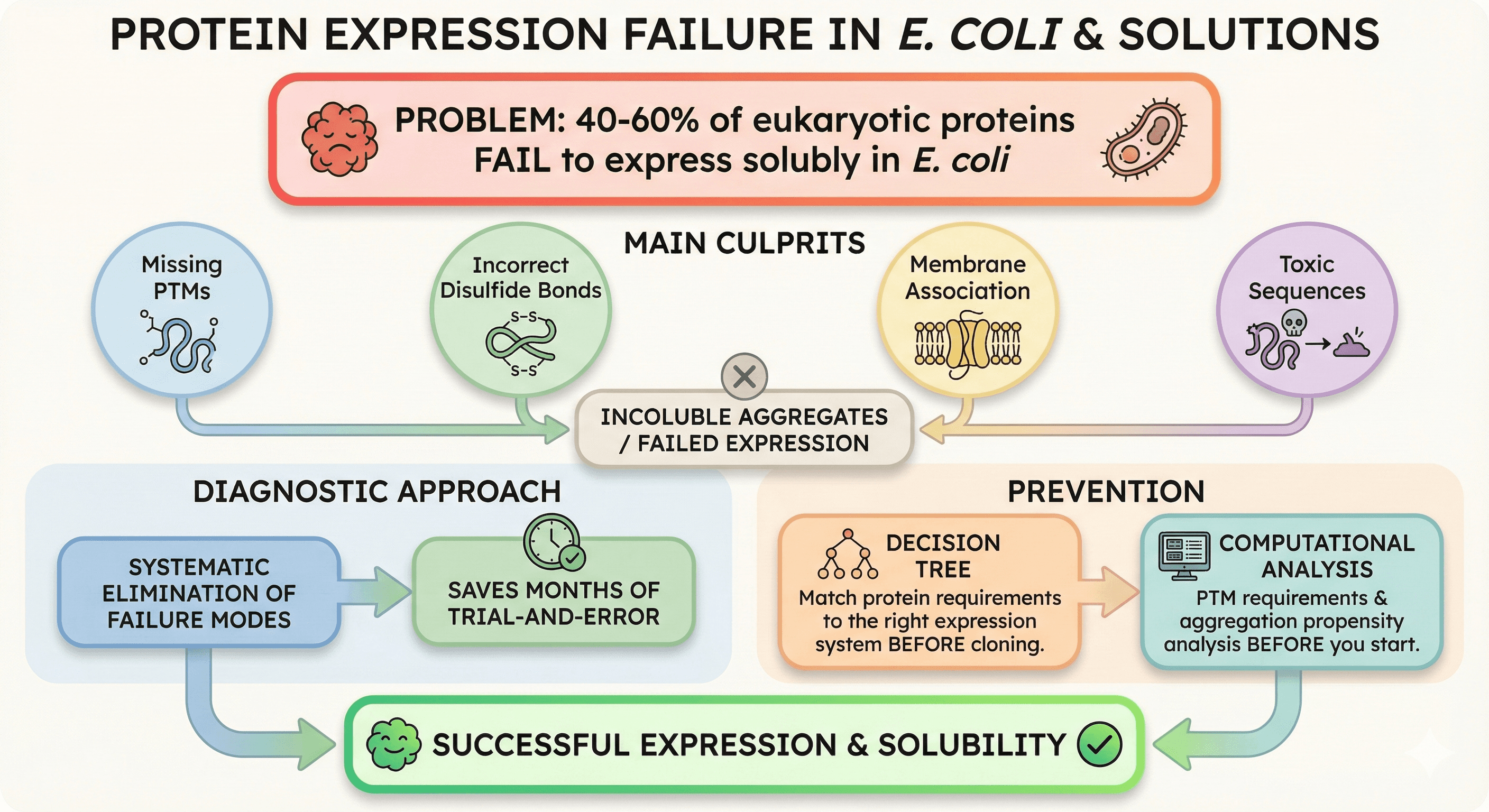
40-60% of eukaryotic proteins fail to express solubly in E. coli
Main culprits: Missing PTMs, incorrect disulfide bonds, membrane association, toxic sequences
Diagnostic approach: Systematic elimination of failure modes saves months of trial-and-error
Decision tree: Match your protein's requirements to the right expression system before cloning
Prevention: Computational analysis of PTM requirements and aggregation propensity before you start
The E. coli Paradox
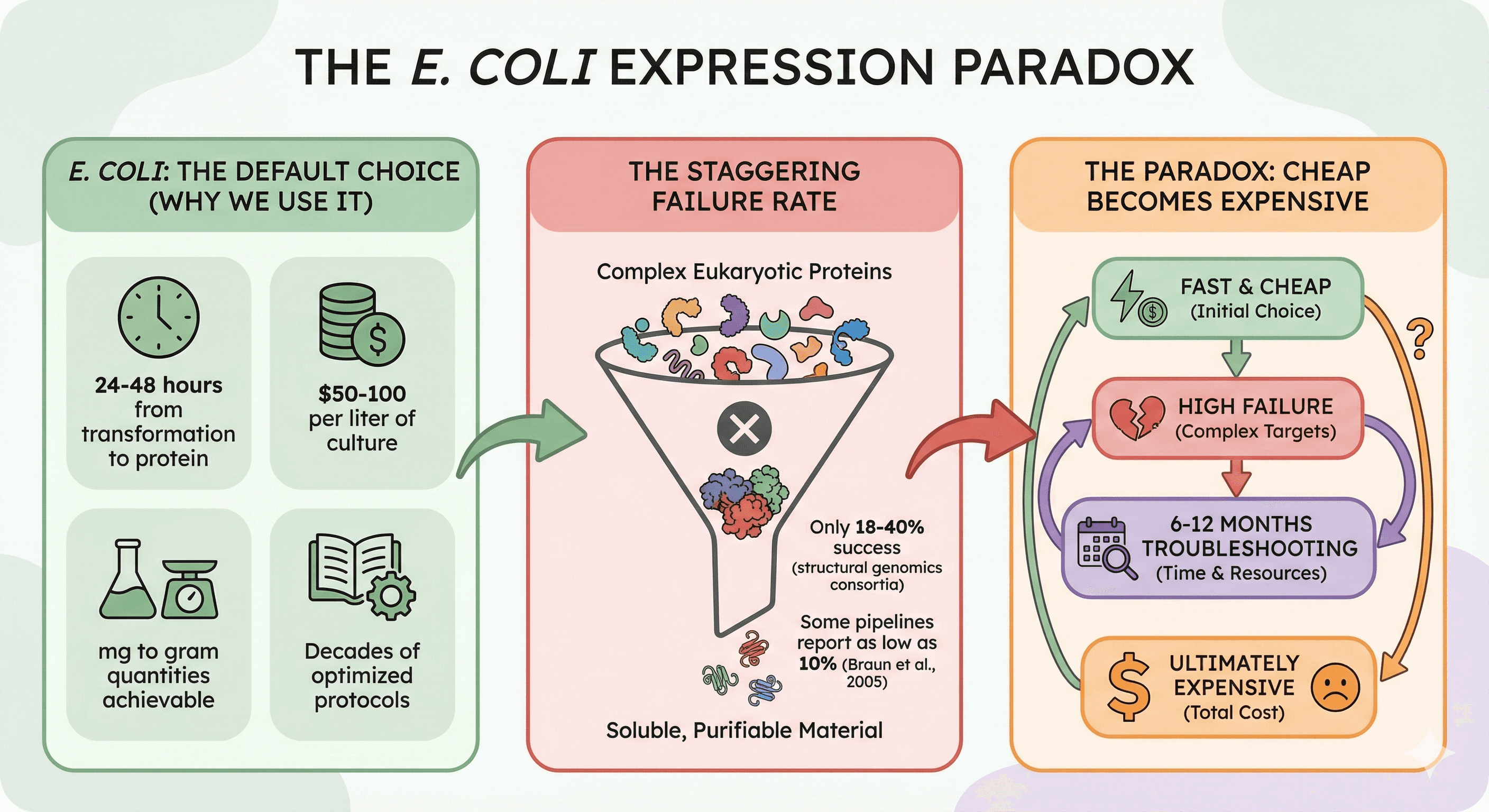
E. coli remains the default choice for recombinant protein expression. And for good reason:
24-48 hours from transformation to protein
$50-100 per liter of culture
mg to gram quantities achievable
Decades of optimized protocols
Yet the failure rate for complex eukaryotic proteins is staggering. Structural genomics consortia report that only 18-40% of human proteins yield soluble, purifiable material from bacterial expression, with some pipelines reporting as low as 10% success for eukaryotic targets (Braun et al., 2005).
The paradox: We keep using E. coli because it's fast and cheap, then spend 6-12 months troubleshooting when it fails. The "cheap" option becomes the expensive one.
The Five Reasons Your Protein Failed
When E. coli expression fails, it's almost always one of these five problems. Diagnosing which one saves you from random troubleshooting.
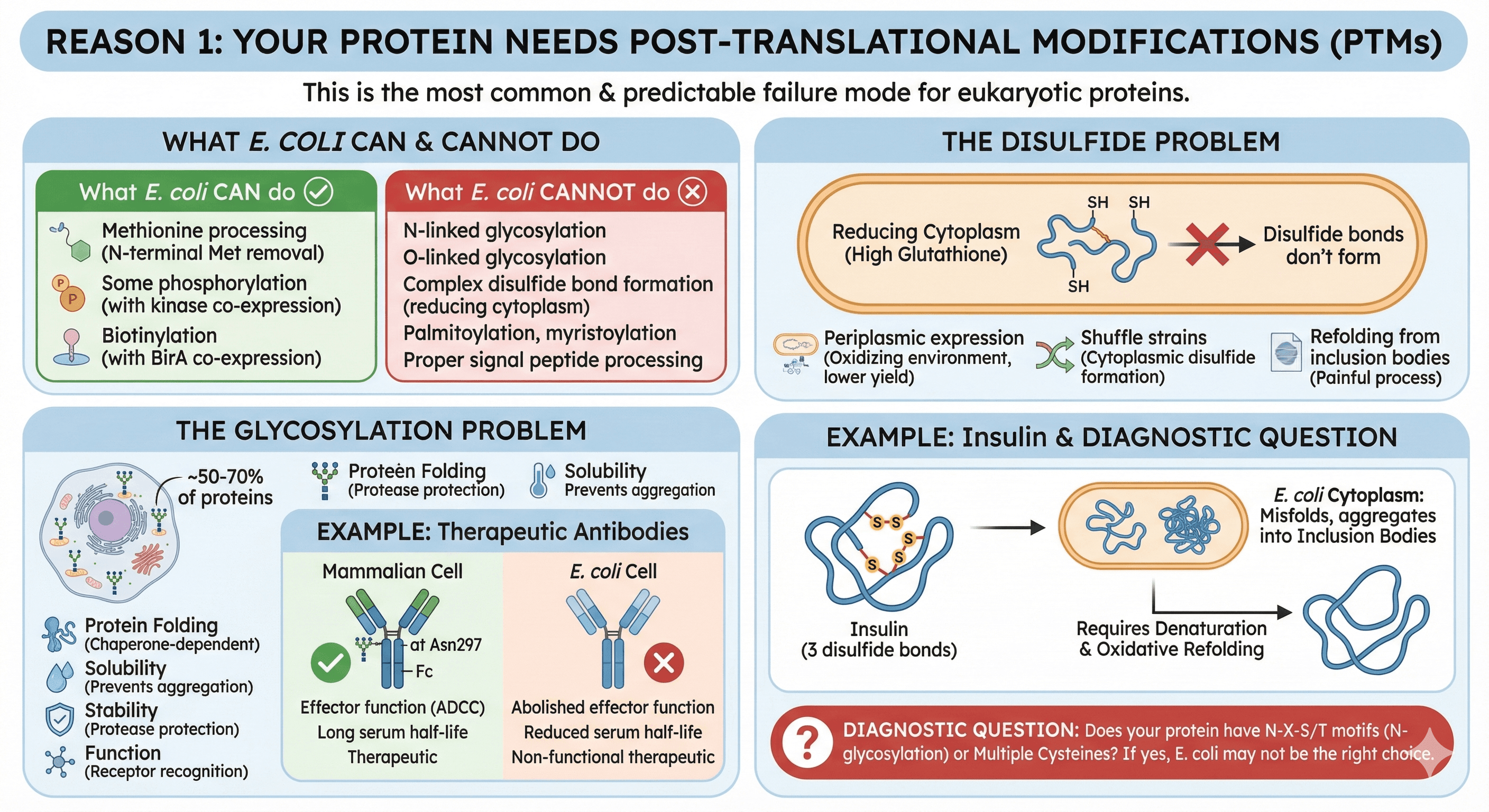
Reason 1: Your Protein Needs Post-Translational Modifications
This is the most common failure mode for eukaryotic proteins, and the most predictable.
What E. coli can do:
Methionine processing (N-terminal Met removal)
Some phosphorylation (if you co-express the kinase)
Biotinylation (with BirA co-expression)
What E. coli cannot do:
N-linked glycosylation
O-linked glycosylation
Complex disulfide bond formation (cytoplasm is reducing)
Palmitoylation, myristoylation
Proper signal peptide processing
The glycosylation problem:
Approximately 50% of human proteins are glycosylated, with some estimates suggesting over 70% of the eukaryotic secretory proteome undergoes glycosylation (Apweiler et al., 1999). Glycans aren't just decorations—they're often essential for:
Protein folding (glycan-dependent chaperones like calnexin)
Solubility (hydrophilic glycans prevent aggregation)
Stability (protection from proteases)
Function (receptor recognition, cell signaling)
Example: Therapeutic antibodies
IgG antibodies require N-glycosylation at Asn297 in the Fc region (Wang et al., 2017). Without it:
Effector function (ADCC) is abolished or significantly reduced
Serum half-life drops dramatically
The protein may still fold, but it's non-functional therapeutically
Express an antibody in E. coli and you get protein. But it's not a therapeutic.
The disulfide problem:
E. coli's cytoplasm is reducing (high glutathione, thioredoxin). Disulfide bonds don't form. Options:
Periplasmic expression (oxidizing environment, but lower yield)
Shuffle strains (cytoplasmic disulfide formation)
Refolding from inclusion bodies (works, but painful)
Example: Insulin
Insulin has 3 disulfide bonds (two interchain and one intrachain) (Baeshen et al., 2014). Express in E. coli cytoplasm:
Protein misfolds immediately
Aggregates into inclusion bodies
Requires denaturation and oxidative refolding
This is doable—the majority of recombinant insulin therapeutics are produced from E. coli inclusion bodies—but it's a specialized process requiring careful refolding optimization, not a quick expression test.
Diagnostic question: Does your protein have N-X-S/T motifs (potential N-glycosylation sites)? Multiple cysteines? If yes, E. coli may not be the right choice.
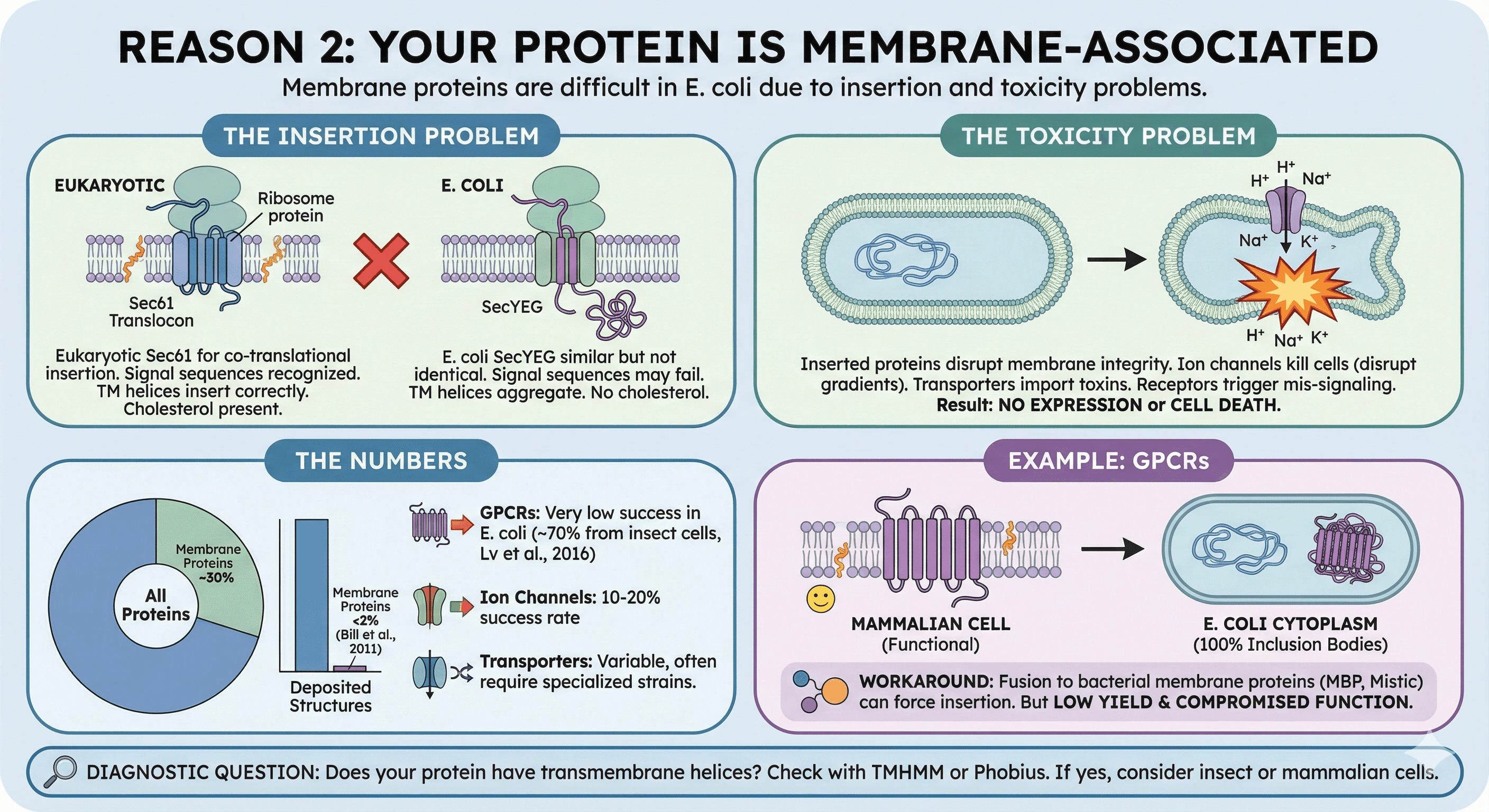
Reason 2: Your Protein Is Membrane-Associated
Membrane proteins are notoriously difficult in E. coli. Not because bacteria lack membranes, but because:
The insertion problem:
Eukaryotic membrane proteins use the Sec61 translocon for co-translational insertion. E. coli uses SecYEG. The machinery is similar but not identical:
Signal sequences may not be recognized
Transmembrane helices may not insert correctly
Lipid composition differs (no cholesterol in bacteria)
The toxicity problem:
Membrane proteins that do insert can disrupt E. coli's membrane integrity:
Ion channels can kill cells by disrupting electrochemical gradients
Transporters can import toxic compounds
Receptors can trigger inappropriate signaling
The result: Either no expression (cells suppress the toxic protein) or cell death (you get no cells to harvest).
The numbers:
Membrane proteins represent ~30% of all proteins yet account for less than 2% of deposited structures (Bill et al., 2011):
GPCRs in E. coli: Very low success rate for functional protein; most GPCR structures (~70%) come from insect cells (Lv et al., 2016)
Ion channels: 10-20% success rate
Transporters: Variable, often require specialized strains
Example: GPCRs
G-protein coupled receptors are 7-transmembrane proteins. In E. coli:
No mammalian membrane insertion machinery
No appropriate lipid environment (needs cholesterol for many GPCRs)
Hydrophobic transmembrane helices aggregate in cytoplasm
Result: 100% inclusion bodies
The workaround: Fusion to bacterial membrane proteins (MBP, Mistic) can sometimes force membrane insertion. But yield is low and function is often compromised.
Diagnostic question: Does your protein have transmembrane helices? Check with TMHMM or Phobius. If yes, consider insect or mammalian cells.
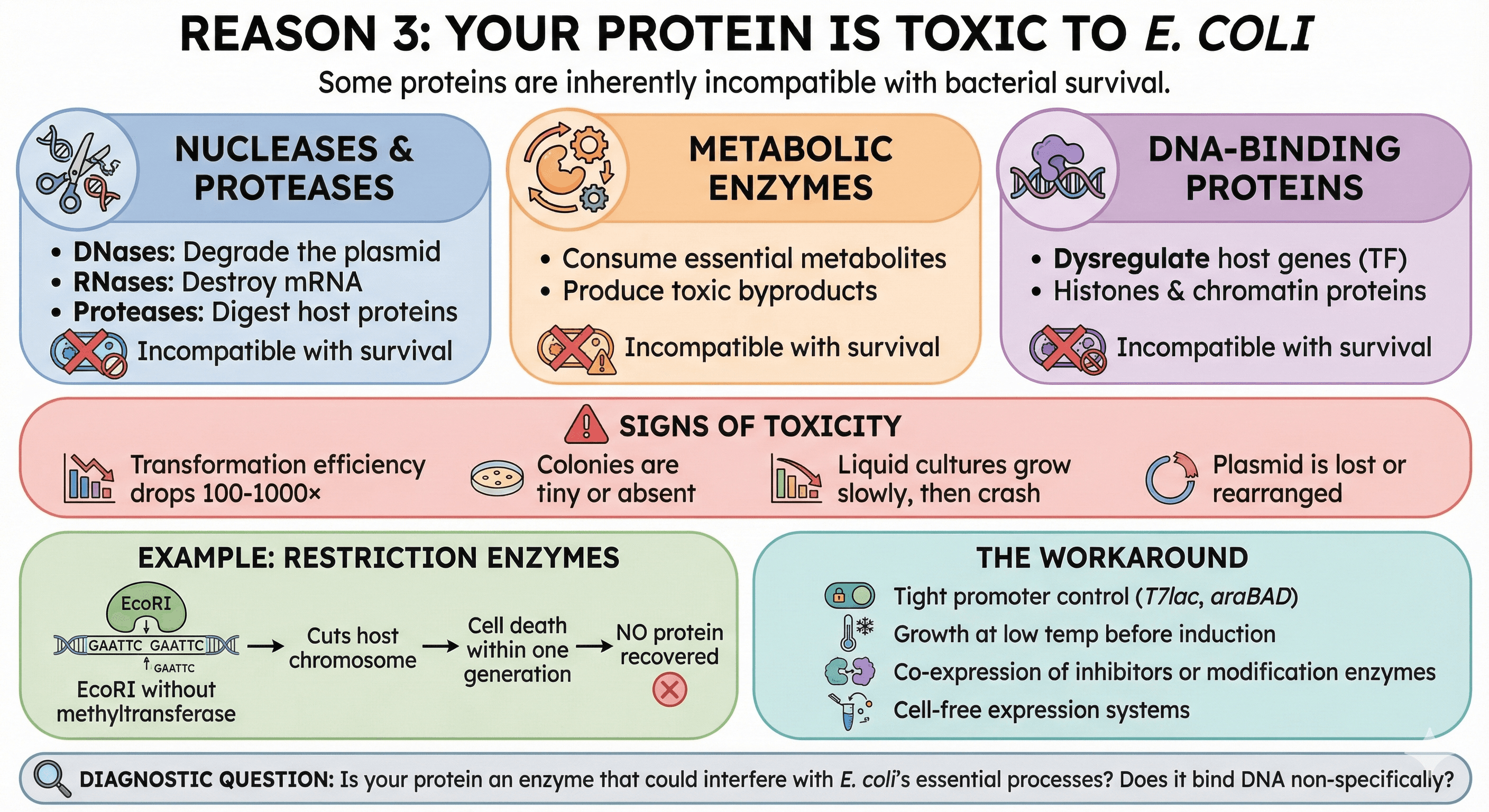
Reason 3: Your Protein Is Toxic to E. coli
Some proteins are inherently incompatible with bacterial survival. This includes:
Nucleases and proteases:
DNases will degrade the plasmid
RNases will destroy mRNA
Proteases will digest host proteins
Metabolic enzymes:
Enzymes that consume essential metabolites
Enzymes that produce toxic byproducts
DNA-binding proteins:
Transcription factors that dysregulate host genes
Histones and chromatin proteins
Signs of toxicity:
Transformation efficiency drops 100-1000×
Colonies are tiny or absent
Liquid cultures grow slowly, then crash
Plasmid is lost or rearranged
Example: Restriction enzymes
EcoRI cuts the recognition sequence GAATTC. E. coli's genome contains hundreds of GAATTC sites. Express EcoRI without its methyltransferase partner, and:
The enzyme cuts the host chromosome
Cell death within one generation
No protein recovered
The workaround:
Tight promoter control (T7lac, araBAD)
Growth at low temperature before induction
Co-expression of inhibitors or modification enzymes
Cell-free expression systems
Diagnostic question: Is your protein an enzyme that could interfere with E. coli's essential processes? Does it bind DNA non-specifically?
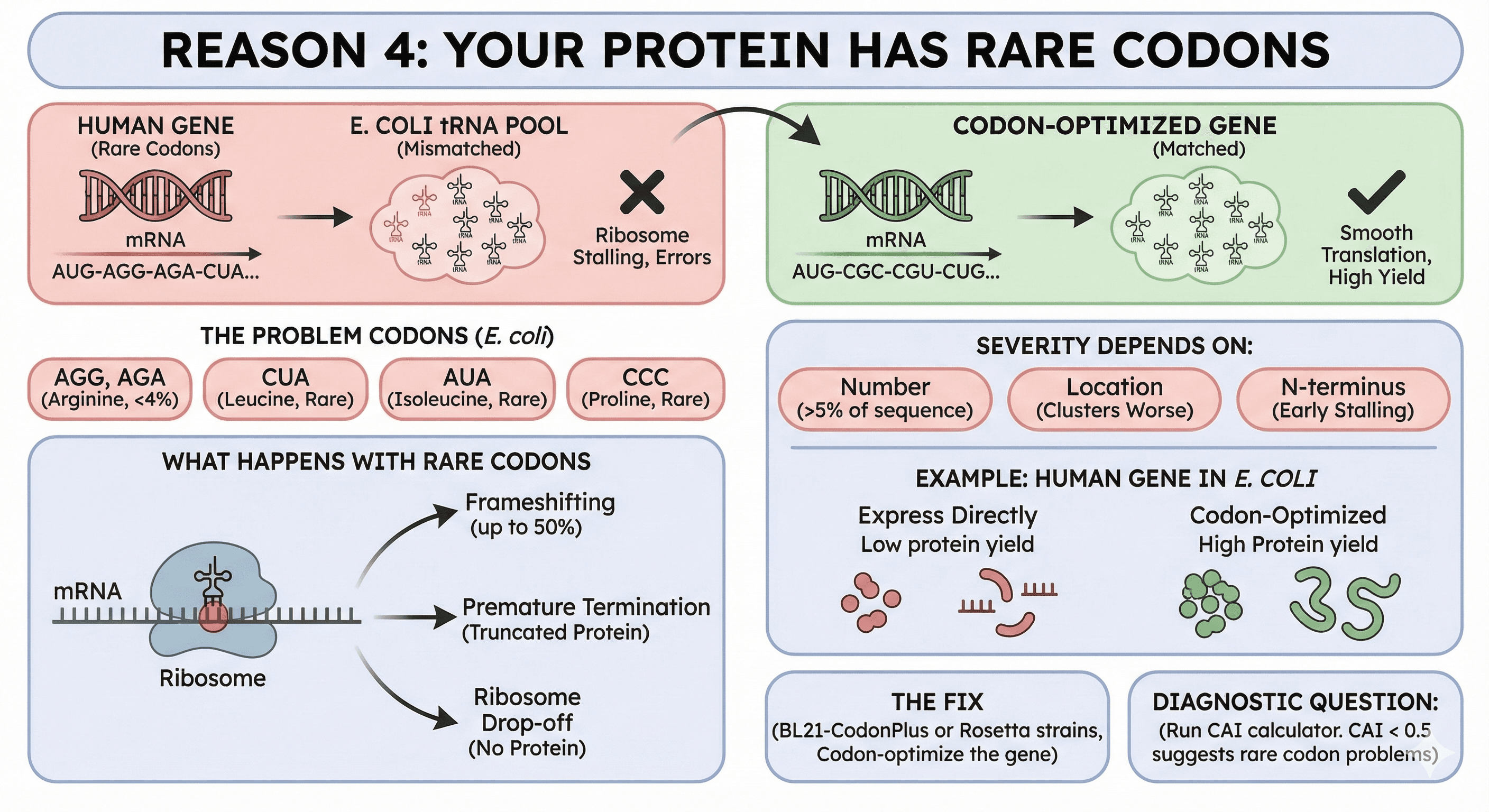
Reason 4: Your Protein Has Rare Codons
E. coli's tRNA pool is optimized for E. coli genes. Human genes use different codon preferences.
The problem codons (Chen & Bhargava, 1994):
AGG, AGA (Arginine): The rarest codons in E. coli, comprising only 2% and 4% of arginine codons respectively
CUA (Leucine): Rare
AUA (Isoleucine): Rare
CCC (Proline): Rare
What happens with rare codons:
Ribosome stalls waiting for the rare tRNA
Stalling causes frameshifting—up to 50% frameshifting at tandem AGG_AGG or AGA_AGA codons (Spanjaard & Van Duin, 1988)
Stalling causes premature termination (truncated protein)
Stalling causes ribosome drop-off (no protein)
The severity depends on:
How many rare codons (>5% of sequence is problematic)
Where they are (clusters are worse than distributed)
Whether they're in the N-terminus (early stalling = no protein)
Example: Human genes in E. coli
A typical human gene might have 10-15% rare codons. Express directly:
Yield drops 10-100× compared to codon-optimized version
Truncation products appear
Full-length protein is often misfolded
The fix:
Use BL21-CodonPlus or Rosetta strains (supply rare tRNAs)
Codon-optimize the gene (change codons, not amino acids)
Both together for difficult cases
Diagnostic question: Run your sequence through a codon adaptation index (CAI) calculator. CAI < 0.5 suggests rare codon problems.
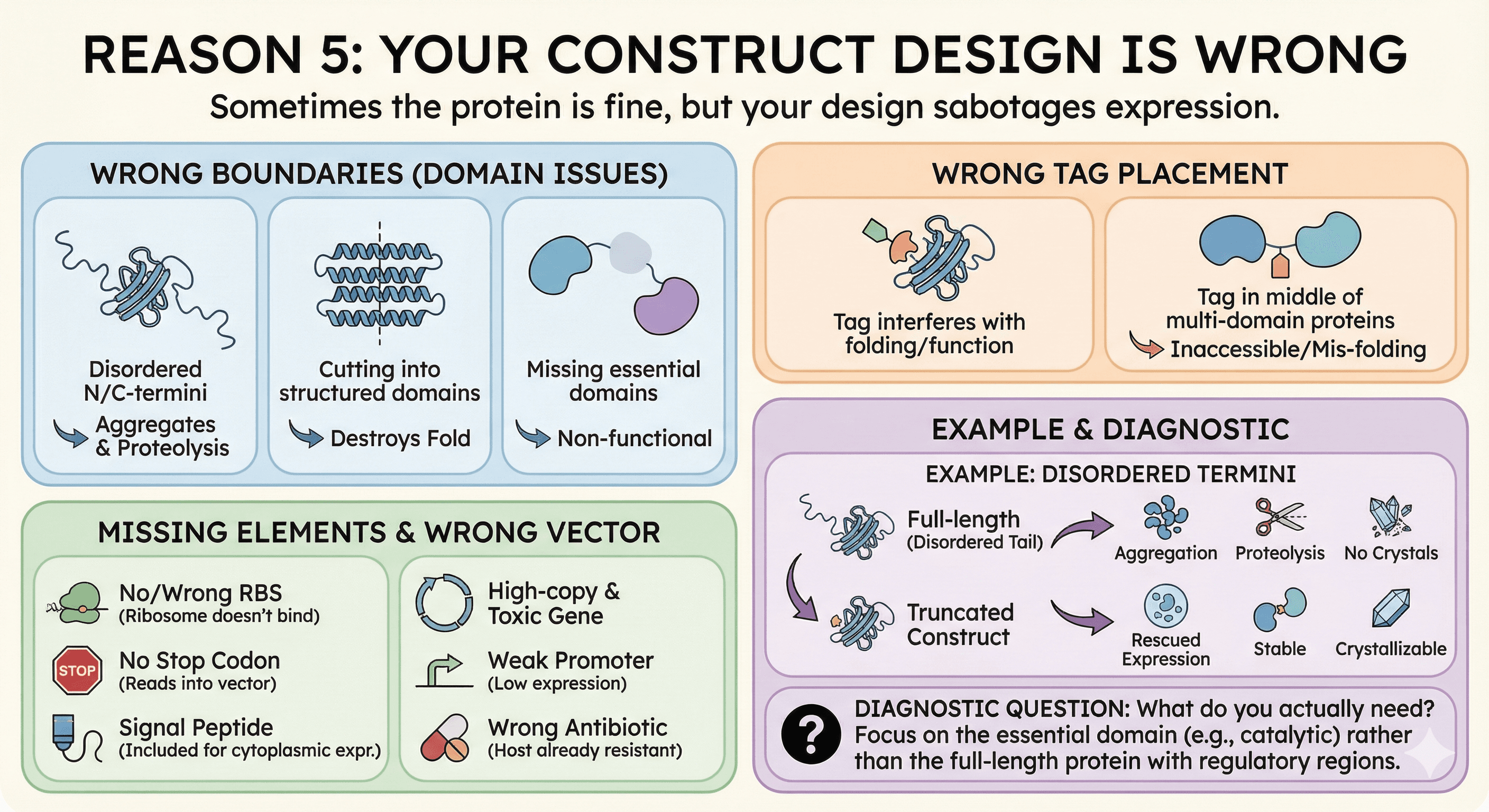
Reason 5: Your Construct Design Is Wrong
Sometimes the protein itself is fine in E. coli, but your construct design sabotages expression.
Common construct problems:
Wrong boundaries:
Including disordered N/C-termini that promote aggregation
Cutting into structured domains (destroys fold)
Missing essential domains (non-functional protein)
Wrong tag placement:
N-terminal tags on proteins that need free N-terminus
Tags that interfere with folding
Tags in the middle of multi-domain proteins
Missing elements:
No ribosome binding site (or wrong spacing)
No stop codon (ribosome reads into vector)
Signal peptide included when cytoplasmic expression intended
Wrong vector:
High-copy plasmid with toxic gene (see Reason 3)
Weak promoter for high-expression needs
Wrong antibiotic resistance (already in host)
Example: Disordered termini
Many proteins have flexible N- or C-terminal tails that are disordered. In the cell, these might be functional (protein-protein interactions, localization). In a test tube:
They promote aggregation
They get cleaved by proteases
They make crystallization impossible
Truncating these regions often rescues expression.
Diagnostic question: What do you actually need? If you want the catalytic domain, express the catalytic domain—not the full-length protein with regulatory regions.
The Decision Tree: Before You Clone
The cheapest experiment is the one you don't run. Before committing to E. coli, run this decision tree:
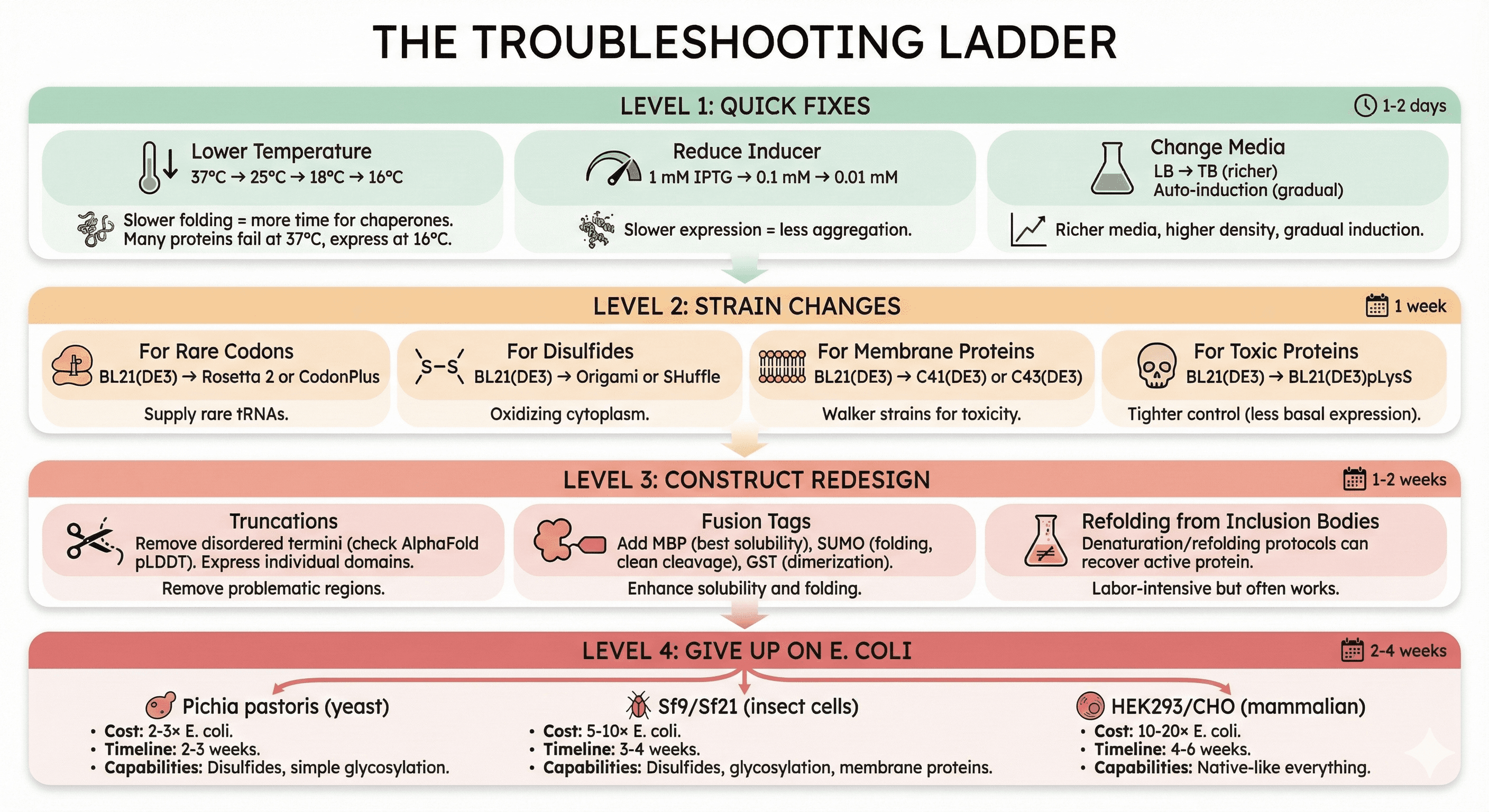
The Troubleshooting Ladder
If you've already tried E. coli and failed, work through this systematically:
Level 1: Quick Fixes (1-2 days)
Lower temperature:
37°C → 25°C → 18°C → 16°C
Slower folding = more time for chaperones
Many proteins that fail at 37°C express at 16°C
Reduce inducer:
1 mM IPTG → 0.1 mM → 0.01 mM
Slower expression = less aggregation
Change media:
LB → TB (richer, higher density)
Auto-induction media (gradual induction)
Level 2: Strain Changes (1 week)
For rare codons:
BL21(DE3) → Rosetta 2 or CodonPlus
For disulfides:
BL21(DE3) → Origami or SHuffle
For membrane proteins:
BL21(DE3) → C41(DE3) or C43(DE3) (Walker strains)
For toxic proteins:
BL21(DE3) → BL21(DE3)pLysS (tighter control)
Level 3: Construct Redesign (1-2 weeks)
Truncations:
Remove disordered termini (check AlphaFold pLDDT)
Express individual domains
Fusion tags:
Add MBP (maltose binding protein) - best for solubility
Add SUMO - improves folding, clean cleavage
Add GST - dimerization can help some proteins
Refolding from inclusion bodies:
Sometimes the protein expresses abundantly but insoluble
Denaturation/refolding protocols can recover active protein
Labor-intensive but often works
Level 4: Give Up on E. coli (2-4 weeks)
If levels 1-3 fail, the protein genuinely needs a eukaryotic system:
Pichia pastoris (yeast):
Cost: 2-3× E. coli
Timeline: 2-3 weeks
Capabilities: Disulfides, simple glycosylation
Sf9/Sf21 (insect cells):
Cost: 5-10× E. coli
Timeline: 3-4 weeks
Capabilities: Disulfides, glycosylation, membrane proteins
HEK293/CHO (mammalian):
Cost: 10-20× E. coli
Timeline: 4-6 weeks
Capabilities: Native-like everything
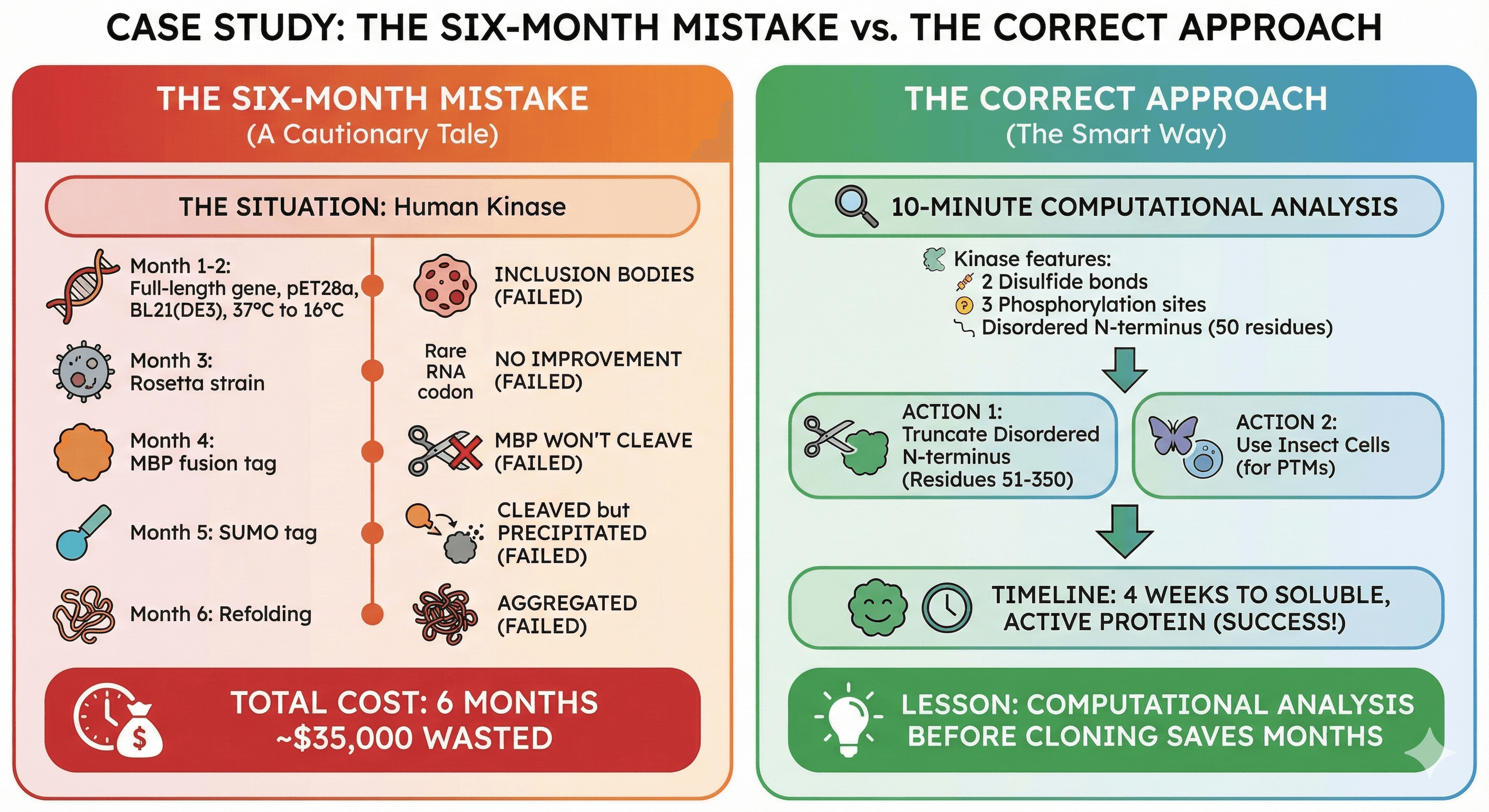
Case Study: The Six-Month Mistake
The Situation
A structural biology lab wanted to crystallize a human kinase. The postdoc:
Cloned full-length gene into pET28a (His-tag)
Expressed in BL21(DE3) at 37°C
Got inclusion bodies
The Six-Month Journey
Months 1-2: Tried every temperature (37°C, 25°C, 18°C, 16°C). Still inclusion bodies.
Month 3: Tried Rosetta strain for rare codons. No improvement.
Month 4: Added MBP fusion tag. Now soluble! But MBP wouldn't cleave off.
Month 5: Tried SUMO tag. Cleaved cleanly. But protein precipitated immediately after cleavage.
Month 6: Tried refolding from inclusion bodies. Got some soluble protein. It was aggregated.
Total cost: 6 months of postdoc time (~$30k), reagents (~$5k), opportunity cost (priceless).
What Should Have Happened
Before cloning, a 10-minute analysis would have revealed:
The kinase has 4 cysteines that form 2 disulfide bonds
It has 3 phosphorylation sites essential for activity
The N-terminal 50 residues are disordered and aggregation-prone
The correct approach:
Truncate disordered N-terminus (residues 51-350)
Use insect cells (disulfides + phosphorylation)
Timeline: 4 weeks to soluble, active protein
Lesson: Computational analysis before cloning saves months.
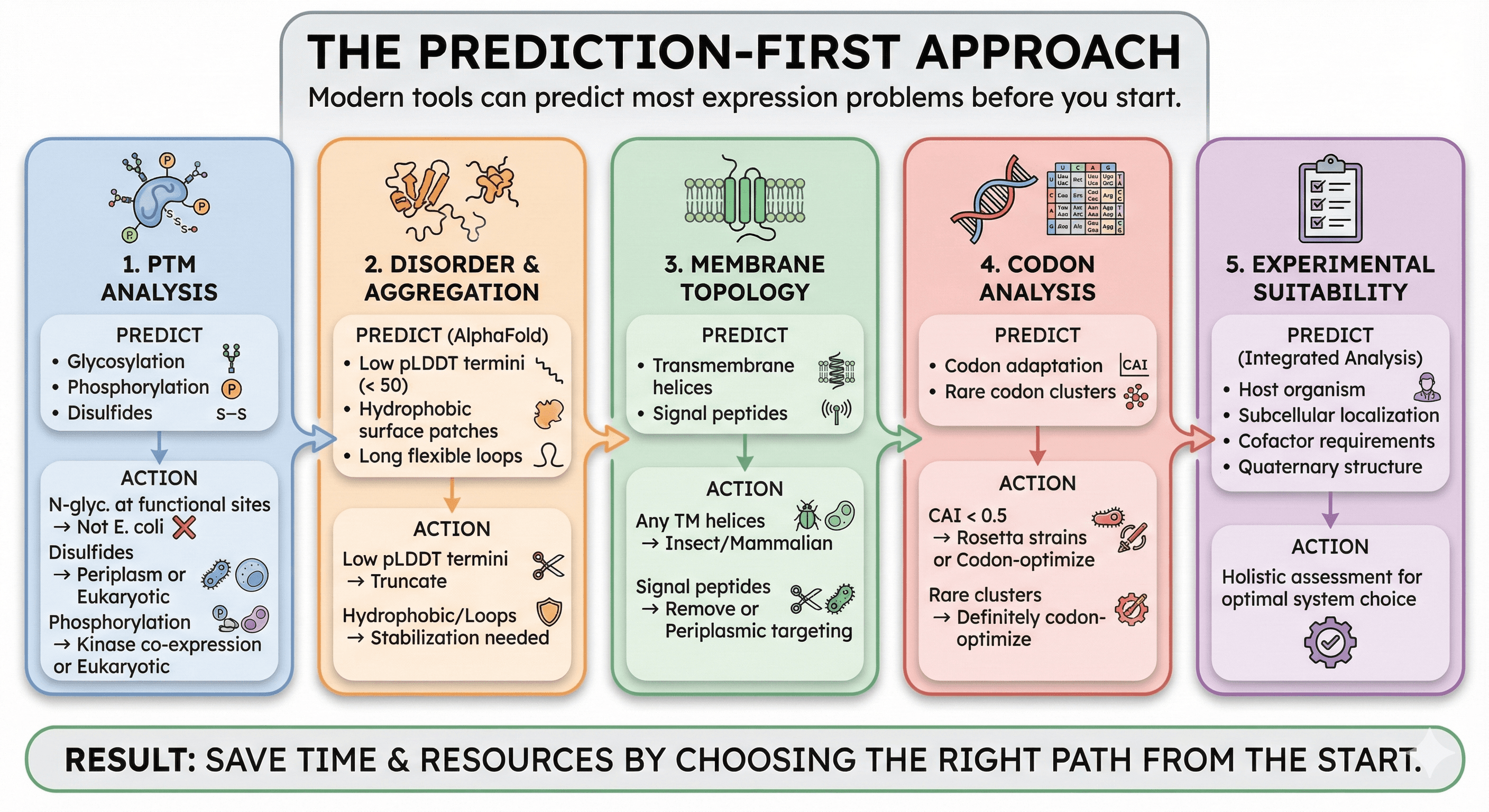
The Prediction-First Approach
Modern tools can predict most expression problems before you start:
1. PTM Analysis
Predict glycosylation, phosphorylation, and other modifications:
If N-glycosylation is predicted at functional sites → not E. coli
If disulfides are predicted → consider periplasm or eukaryotic
If phosphorylation is required for activity → need kinase co-expression or eukaryotic
2. Disorder and Aggregation
Check AlphaFold structure for:
Low pLDDT regions (< 50) at termini → candidate for truncation
Hydrophobic patches on surface → aggregation risk
Long flexible loops → may need stabilization
3. Membrane Topology
Predict transmembrane helices:
Any TM helices → consider insect/mammalian
Signal peptides → may need removal or periplasmic targeting
4. Codon Analysis
Check codon adaptation:
CAI < 0.5 → use Rosetta strains or codon-optimize
Clusters of rare codons → definitely codon-optimize
5. Experimental Suitability
Integrated analysis that considers:
Host organism association
Subcellular localization
Cofactor requirements
Quaternary structure
The Bottom Line
E. coli expression failure isn't random bad luck. It's a predictable consequence of mismatched requirements:
Your Protein Needs | E. coli Provides | Result |
|---|---|---|
Glycosylation | Nothing | Misfolding |
Disulfide bonds | Reducing cytoplasm | Aggregation |
Membrane insertion | Bacterial translocon | Inclusion bodies |
Rare tRNAs | Standard tRNA pool | Truncation |
Complex folding | Fast translation | Misfolding |
The old approach: Clone → Express → Fail → Troubleshoot for months
The new approach: Analyze → Predict requirements → Choose system → Express → Succeed
The difference between a "difficult protein" and a "successful expression" is often just choosing the right system before you start—not after six months of failure.
Matching Your Protein to the Right System
For researchers dealing with expression failures, platforms like Orbion can predict PTM requirements, aggregation hotspots, and expression system compatibility before you clone. The analysis takes minutes; the troubleshooting it prevents takes months.
What Orbion provides:
PTM prediction (glycosylation, phosphorylation, disulfides)
Experimental suitability assessment (host organism, subcellular location, cofactors)
Aggregation propensity mapping
Expression system recommendations based on your protein's specific requirements
The goal isn't to avoid E. coli—it's the best system when it works. The goal is to know when it won't work, before you waste months discovering that the hard way.
References
Braun P, et al. (2005). Structural genomics of human proteins – target selection and generation of a public catalogue of expression clones. Microbial Cell Factories, 4:21. PMC1250228
Apweiler R, et al. (1999). On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochimica et Biophysica Acta, 1473(1):4-8.
Wang W, et al. (2017). Glycosylation engineering of therapeutic IgG antibodies: challenges for the safety, functionality and efficacy. Protein & Cell, 9(1):16-25. Link
Baeshen MN, et al. (2021). Downstream processing of recombinant human insulin and its analogues production from E. coli inclusion bodies. Bioresources and Bioprocessing, 8:78. PMC8313369
Bill RM, et al. (2011). High-throughput expression and purification of membrane proteins. Journal of Structural Biology, 172(1):73-82. PMC2933282
Lv X, et al. (2016). Expression and purification of recombinant G protein-coupled receptors: A review. Protein Expression and Purification, 123:1-6. PMC6983937
Chen GT & Bhargava MM. (1994). Role of the AGA/AGG codons, the rarest codons in global gene expression in Escherichia coli. Genes & Development, 8(21):2641-52. Link
Spanjaard RA & Van Duin J. (2005). Expression levels influence ribosomal frameshifting at the tandem rare arginine codons AGG_AGG and AGA_AGA in Escherichia coli. Journal of Bacteriology, 187(12):4023-4032. PMC1151738

