Blog
Fixing Cryo-EM Sample Problems: Optimization Strategies & Workflows
Jan 7, 2026
We've diagnosed the 5 cryo-EM sample killers: preferred orientation, aggregation, denaturation, heterogeneity, and ice quality issues. Now comes the critical question: How do you fix it?
Nw, we'll cover systematic solutions, optimization workflows, modern AI-driven prediction, and a real-world case study of rescuing a failed ion channel project.
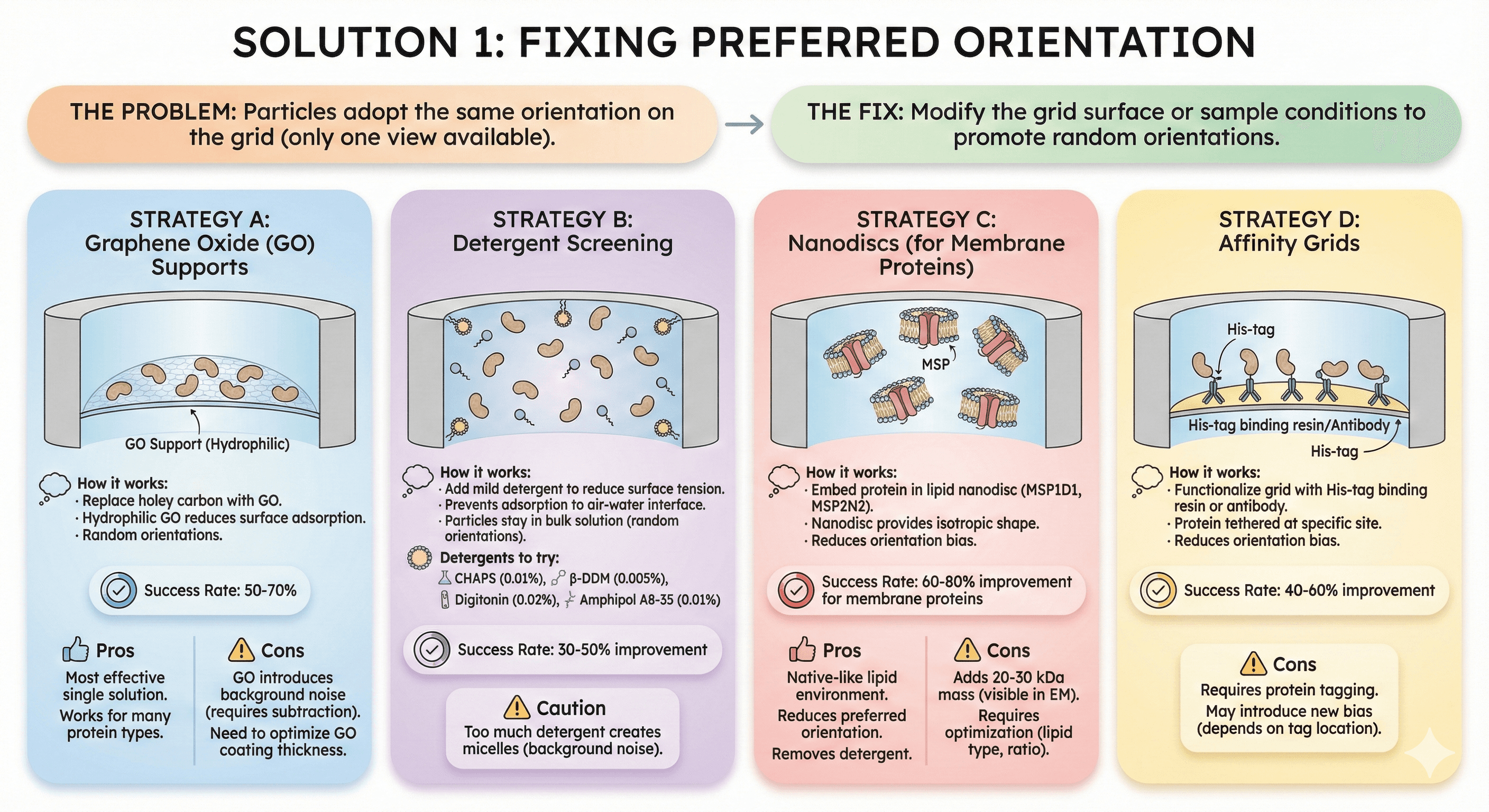
Solution 1: Fixing Preferred Orientation
The problem: Particles adopt the same orientation on the grid (only one view available).
The fix: Modify the grid surface or sample conditions to promote random orientations.
Strategy A: Graphene Oxide (GO) Supports
How it works:
Replace holey carbon with graphene oxide
GO is hydrophilic (reduces surface adsorption)
Particles adopt random orientations
Success rate: 50-70% of preferred orientation problems solved
Pros:
Most effective single solution
Works for many protein types
Cons:
GO introduces background noise (requires subtraction)
Need to optimize GO coating thickness
Strategy B: Detergent Screening
How it works:
Add mild detergent to reduce surface tension
Prevents adsorption to air-water interface
Particles stay in bulk solution (random orientations)
Detergents to try:
CHAPS (0.01%)
β-DDM (0.005%)
Digitonin (0.02%)
Amphipol A8-35 (0.01%)
Success rate: 30-50% improvement
Caution: Too much detergent creates micelles (background noise)
Strategy C: Nanodiscs (for Membrane Proteins)
How it works:
Embed protein in lipid nanodisc (MSP1D1, MSP2N2)
Nanodisc provides isotropic shape
Reduces orientation bias
Success rate: 60-80% improvement for membrane proteins
Pros:
Native-like lipid environment
Reduces preferred orientation
Removes detergent
Cons:
Adds 20-30 kDa mass (visible in EM)
Requires optimization (lipid type, ratio)
Strategy D: Affinity Grids
How it works:
Functionalize grid with His-tag binding resin or antibody
Protein tethered at specific site
Reduces orientation bias
Success rate: 40-60% improvement
Cons:
Requires protein tagging
May introduce new bias (depends on tag location)
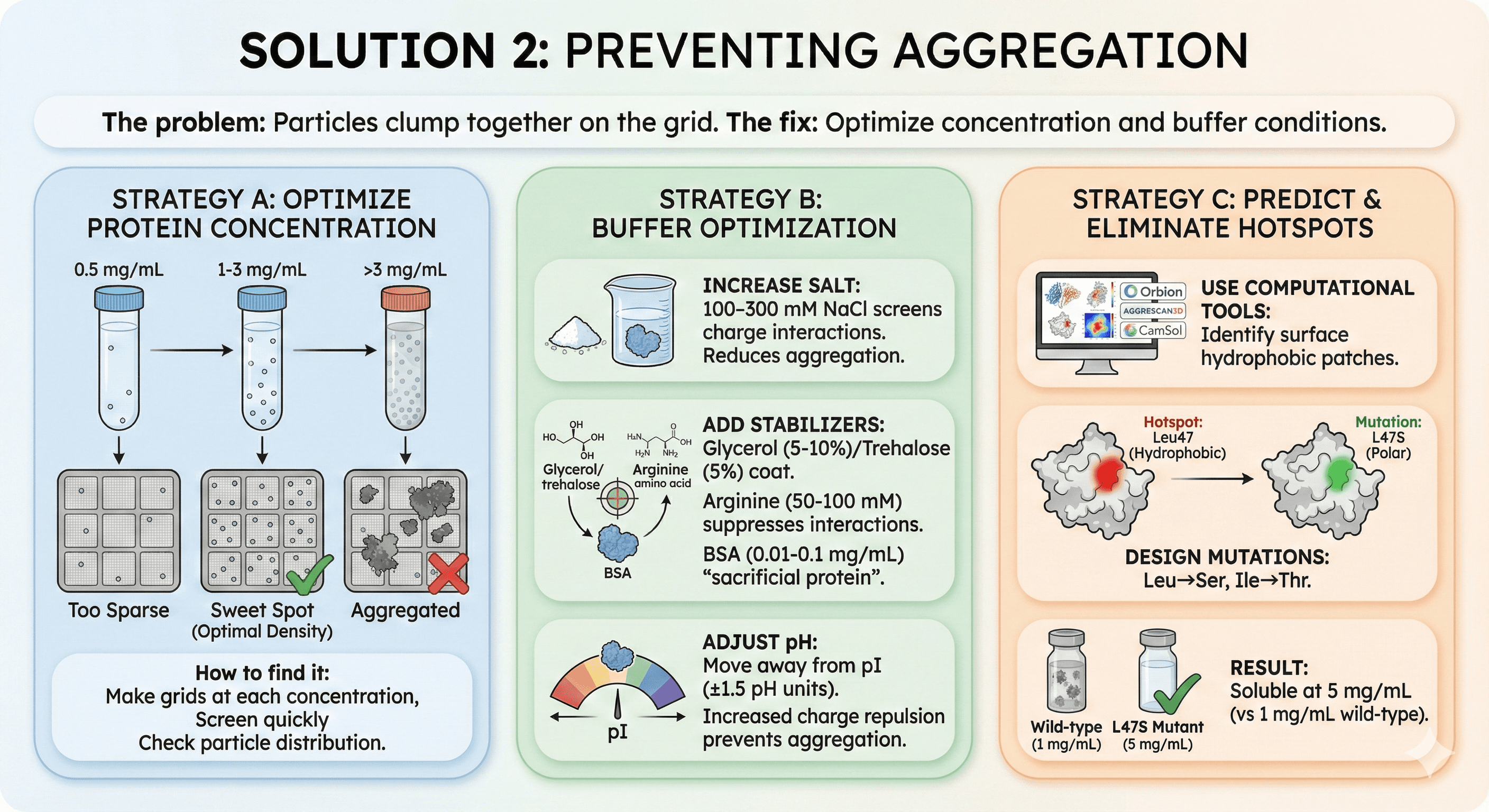
Solution 2: Preventing Aggregation
The problem: Particles clump together on the grid.
The fix: Optimize concentration and buffer conditions.
Strategy A: Optimize Protein Concentration
Test range: 0.5, 1, 2, 3 mg/mL
Sweet spot: High enough for particle density, low enough to avoid aggregation Typical optimal: 1-3 mg/mL for most proteins
How to find it:
Make grids at each concentration
Screen quickly
Check particle distribution
Strategy B: Buffer Optimization
Increase salt:
100-300 mM NaCl screens charge interactions
Reduces aggregation
Add stabilizers:
Glycerol (5-10%) or trehalose (5%)
Arginine (50-100 mM) suppresses protein-protein interactions
BSA (0.01-0.1 mg/mL) as "sacrificial protein"
Adjust pH:
Move away from pI (±1.5 pH units)
Increased charge repulsion prevents aggregation
Strategy C: Predict and Eliminate Aggregation Hotspots
Use computational tools:
Orbion, AGGRESCAN3D, CamSol
Identify surface hydrophobic patches
Design mutations (Leu→Ser, Ile→Thr)
Example:
Hotspot: Leu47 on surface
Mutation: L47S
Result: Soluble at 5 mg/mL (vs 1 mg/mL wild-type)
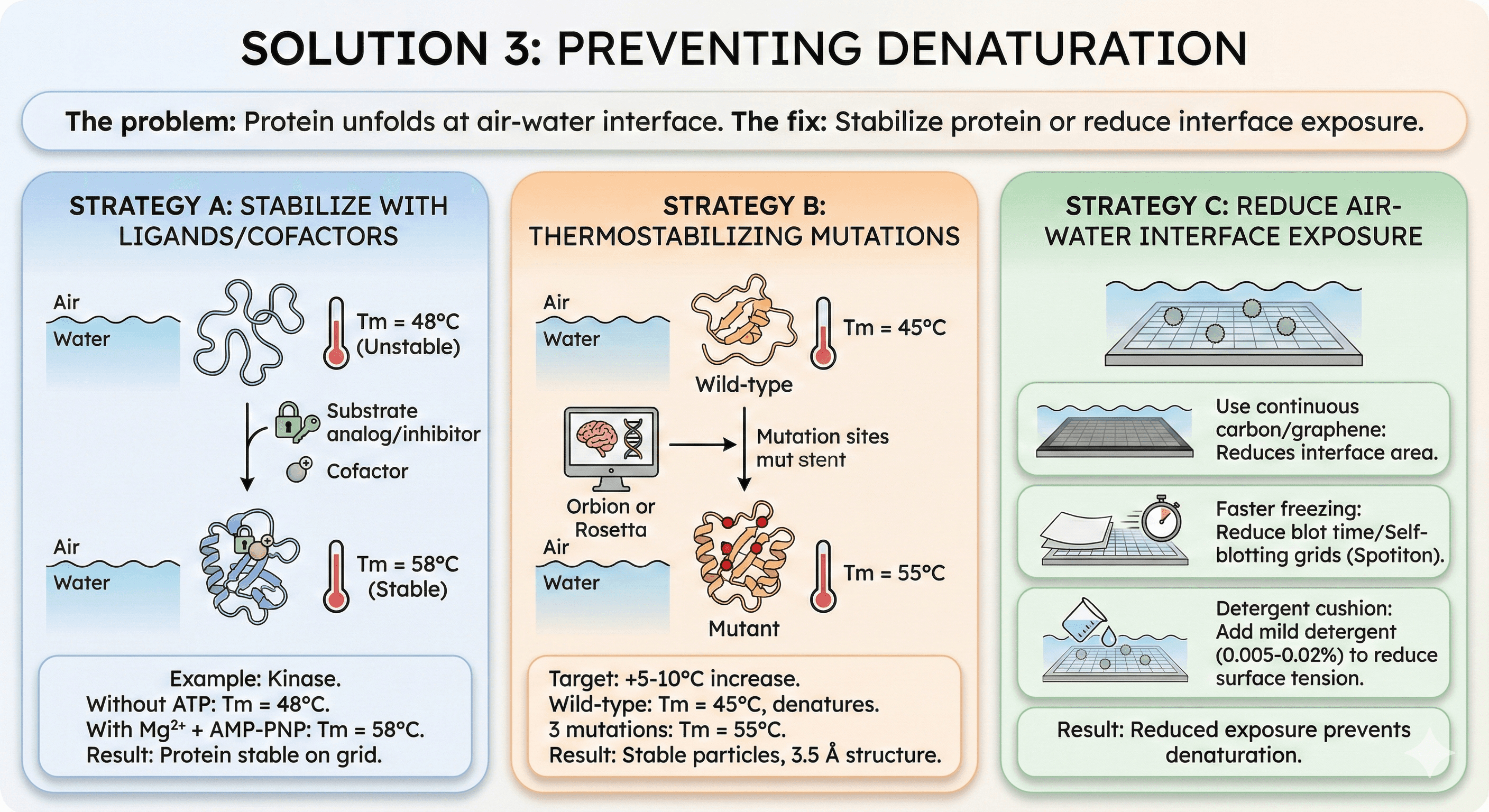
Solution 3: Preventing Denaturation
The problem: Protein unfolds at air-water interface.
The fix: Stabilize protein or reduce interface exposure.
Strategy A: Stabilize with Ligands/Cofactors
Add saturating ligand:
Substrate analog, inhibitor, or cofactor
Stabilizes conformation
Increases Tm
Example: Kinase
Without ATP: Tm = 48°C
With Mg²⁺ + AMP-PNP: Tm = 58°C
Result: Protein stable on grid
Strategy B: Thermostabilizing Mutations
Design mutations to increase Tm:
Target: +5-10°C increase
Use Orbion or Rosetta
Stable proteins resist denaturation
Example:
Wild-type: Tm = 45°C, denatures on grid
3 mutations (predicted by Orbion): Tm = 55°C
Result: Stable particles, 3.5 Å structure
Strategy C: Reduce Air-Water Interface Exposure
Use continuous carbon:
Reduces interface area
Or graphene/graphene oxide
Faster freezing:
Reduce blot time (less time at interface)
Self-blotting grids (Spotiton)
Detergent cushion:
Add mild detergent (0.005-0.02%)
Reduces surface tension
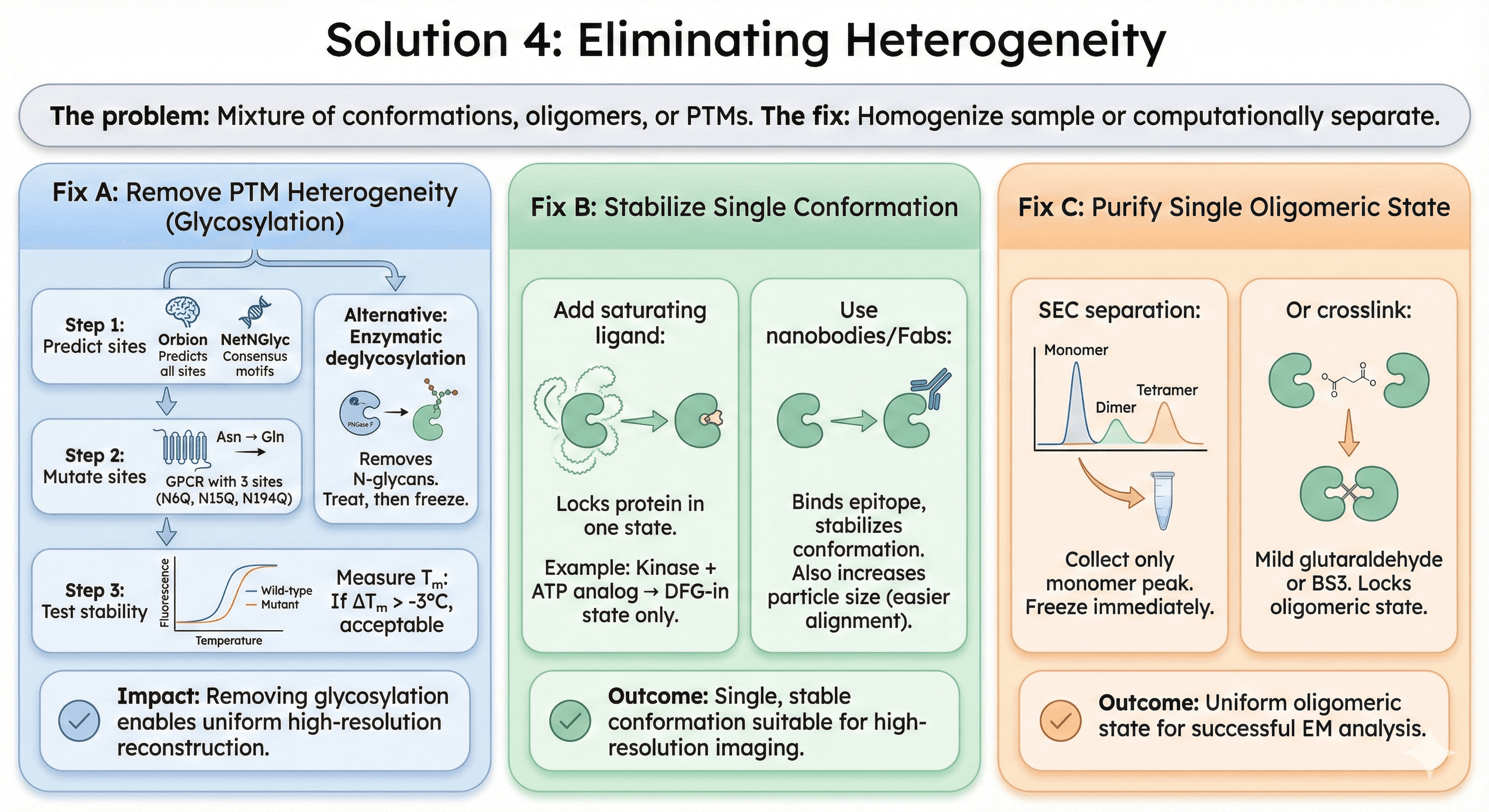
Solution 4: Eliminating Heterogeneity
The problem: Mixture of conformations, oligomers, or PTMs.
The fix: Homogenize sample or computationally separate.
Fix A: Remove PTM Heterogeneity
For glycosylation:
Step 1: Predict sites
Use Orbion (predicts all glycosylation sites)
Or NetNGlyc (free, consensus motifs)
Step 2: Mutate sites
Asn → Gln (blocks N-glycosylation)
Example: GPCR with 3 sites (N6Q, N15Q, N194Q)
Step 3: Test stability
Measure Tm: If ΔTm > -3°C, acceptable
Alternative: Enzymatic deglycosylation
PNGase F removes N-glycans
Treat protein, then freeze for EM
Impact: Removing glycosylation enables uniform high-resolution reconstruction
Fix B: Stabilize Single Conformation
Add saturating ligand:
Locks protein in one state
Example: Kinase + ATP analog → DFG-in state only
Use nanobodies/Fabs:
Binds epitope, stabilizes conformation
Also increases particle size (easier alignment)
Fix C: Purify Single Oligomeric State
SEC separation:
Collect only monomer peak
Freeze immediately
Or crosslink:
Mild glutaraldehyde or BS3
Locks oligomeric state
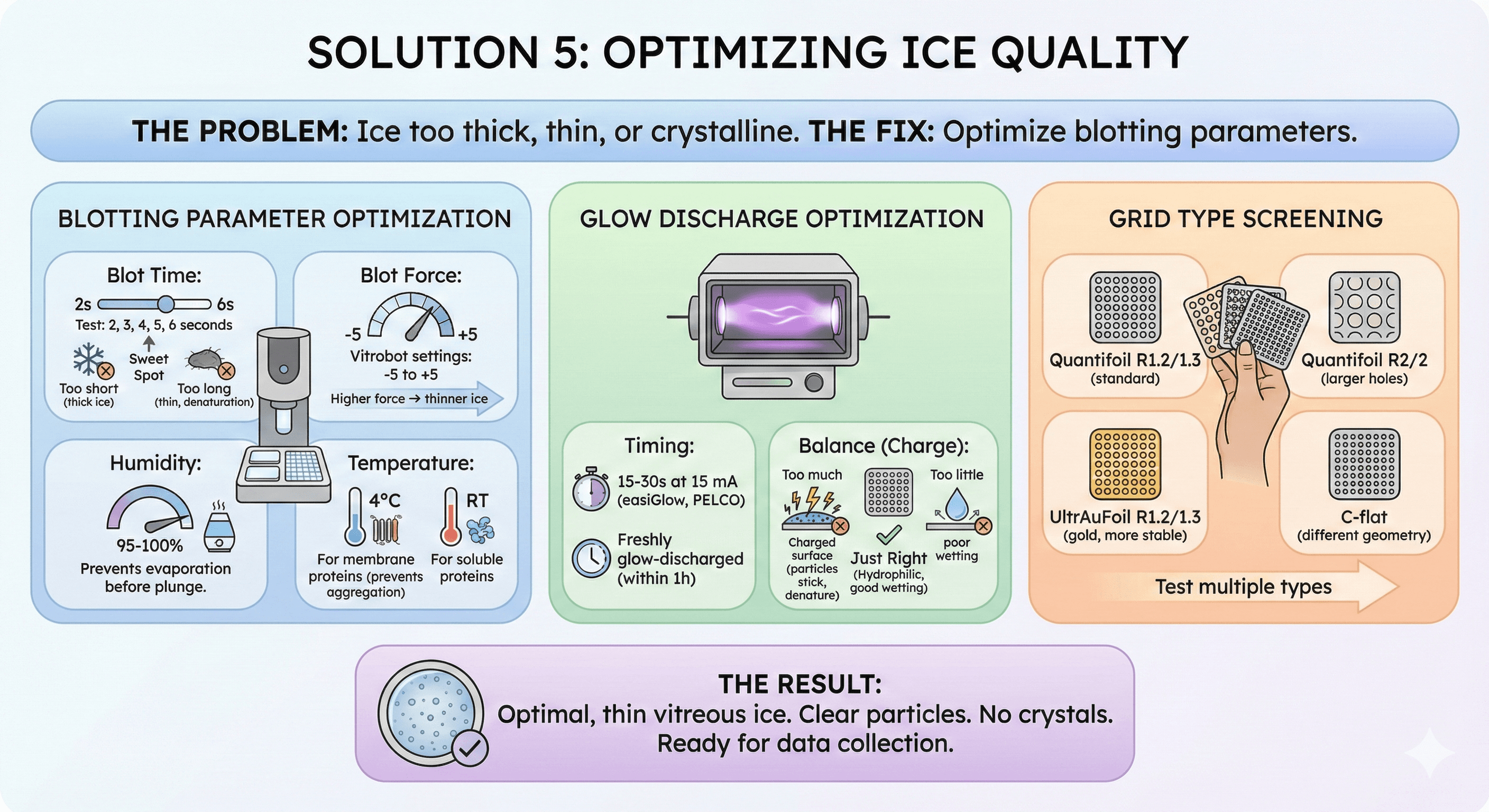
Solution 5: Optimizing Ice Quality
The problem: Ice too thick, thin, or crystalline.
The fix: Optimize blotting parameters.
Blotting Parameter Optimization
Blot time:
Test: 2, 3, 4, 5, 6 seconds
Too short → thick ice
Too long → thin ice, denaturation
Blot force:
Vitrobot settings: -5 to +5
Higher force → thinner ice
Humidity:
95-100% prevents evaporation before plunge
Temperature:
4°C for membrane proteins (prevents aggregation)
Room temperature for soluble proteins
Glow Discharge Optimization
Timing:
15-30 seconds at 15 mA (easiGlow, PELCO)
Freshly glow-discharged (within 1 hour)
Too much:
Charged surface (particles stick, denature)
Too little:
Hydrophobic surface (poor wetting)
Grid Type Screening
Test multiple types:
Quantifoil R1.2/1.3 (standard)
Quantifoil R2/2 (larger holes)
UltrAuFoil R1.2/1.3 (gold, more stable)
C-flat (different geometry)
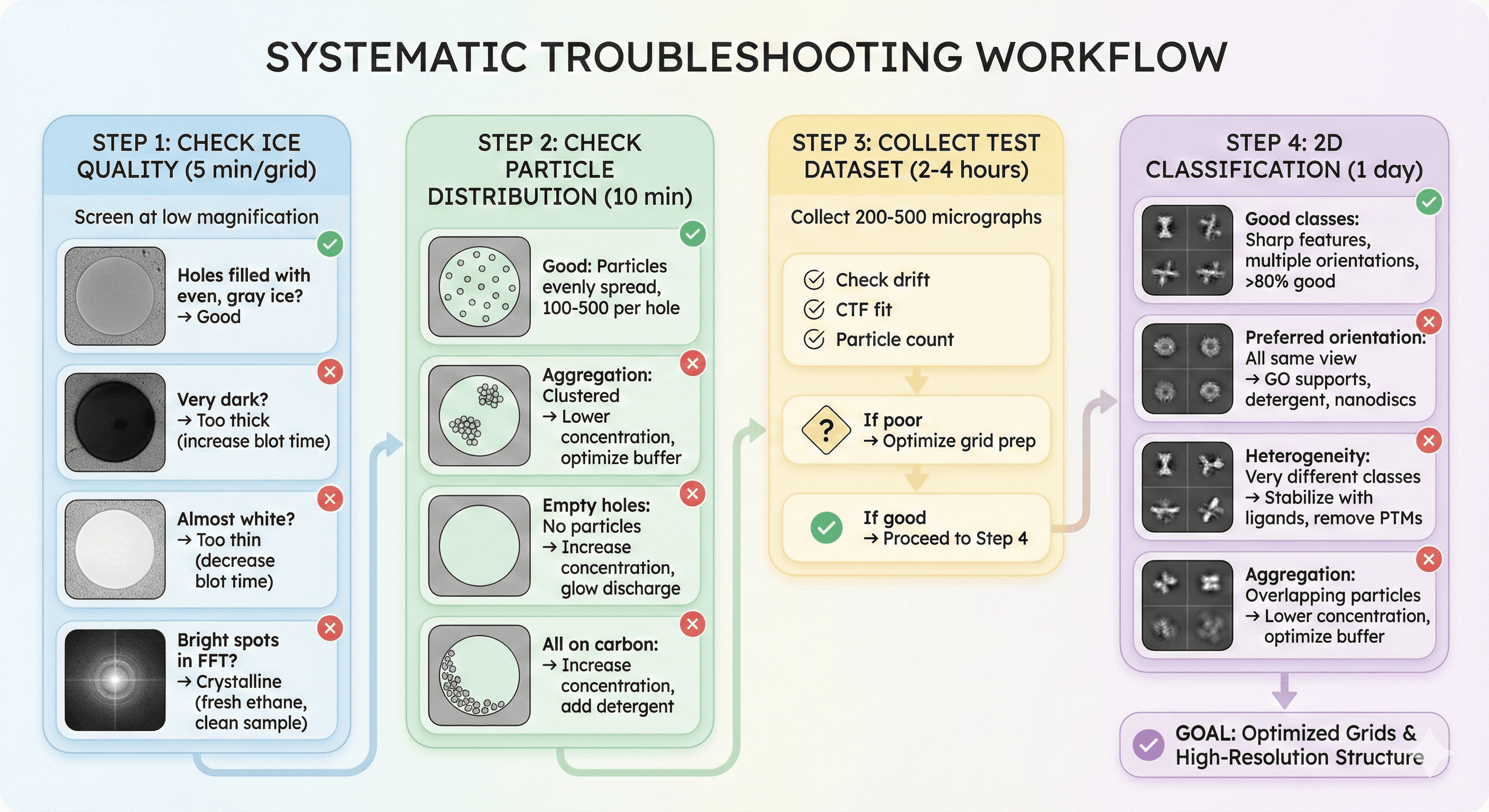
Systematic Troubleshooting Workflow
Step 1: Check Ice Quality (5 min/grid)
Screen at low magnification:
Holes filled with even, gray ice? → Good
Very dark? → Too thick (increase blot time)
Almost white? → Too thin (decrease blot time)
Bright spots in FFT? → Crystalline (fresh ethane, clean sample)
Step 2: Check Particle Distribution (10 min)
Good: Particles evenly spread, 100-500 per hole Aggregation: Clustered → Lower concentration, optimize buffer Empty holes: No particles → Increase concentration, glow discharge All on carbon: → Increase concentration, add detergent
Step 3: Collect Test Dataset (2-4 hours)
Collect 200-500 micrographs:
Check drift, CTF fit, particle count
If poor → Optimize grid prep
Step 4: 2D Classification (1 day)
Good classes: Sharp features, multiple orientations, >80% good Preferred orientation: All same view → GO supports, detergent, nanodiscs Heterogeneity: Very different classes → Stabilize with ligands, remove PTMs Aggregation: Overlapping particles → Lower concentration, optimize buffer
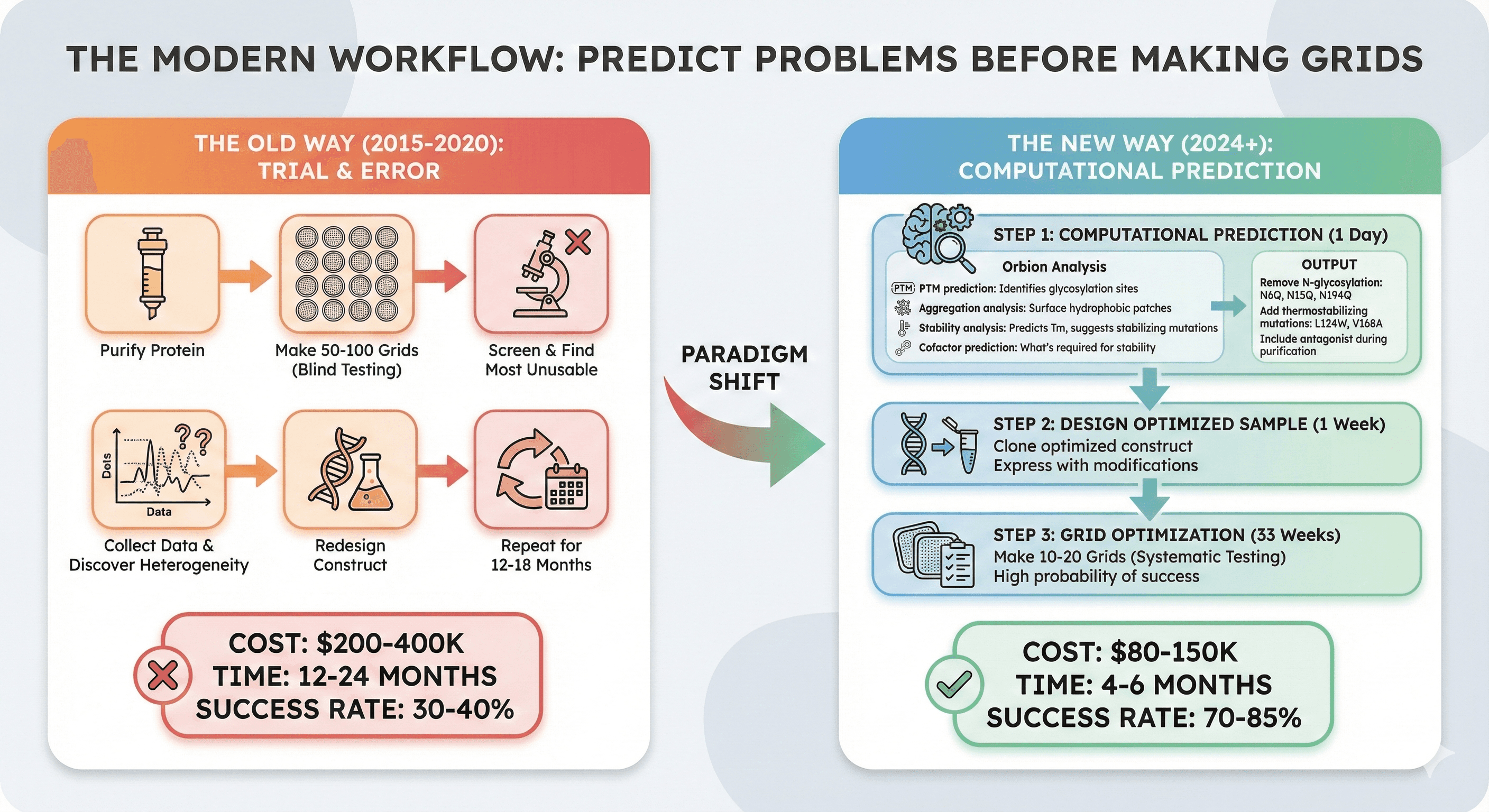
The Modern Workflow: Predict Problems Before Making Grids
The old way (2015-2020):
Purify protein
Make 50-100 grids (blind testing)
Screen → Find most unusable
Collect data → Discover heterogeneity
Redesign construct
Repeat for 12-18 months
Cost: $200-400K, 12-24 months
The new way (2024+):
Step 1: Computational Prediction (1 day)
Orbion analysis:
PTM prediction: Identifies glycosylation sites
Aggregation analysis: Surface hydrophobic patches
Stability analysis: Predicts Tm, suggests stabilizing mutations
Cofactor prediction: What's required for stability
Output:
"Remove N-glycosylation: N6Q, N15Q, N194Q"
"Add thermostabilizing mutations: L124W, V168A"
"Include antagonist during purification"
Step 2: Design Optimized Sample (1 week)
Clone optimized construct
Express with modifications
Step 3: Grid Optimization (3 weeks)
Make 10-20 grids (systematic testing)
High probability of success
Cost: $80-150K, 4-6 months
Success rate: 70-85% (vs 30-40% traditional)
Case Study: Rescuing a Failed Ion Channel
The Challenge
Target: Pentameric ligand-gated ion channel
Initial attempt (2018-2019):
Construct: Full-length
Expression: Sf9 cells, detergent-solubilized
Grids: 80 made
Data: 3 sessions ($150K)
Result: Cannot refine beyond 6 Å
Diagnosis: Severe preferred orientation (>95% top-down), glycosylation heterogeneity
The Rescue (with Orbion)
Step 1: PTM analysis
Orbion predicts 4 N-glycosylation sites per subunit (20 total)
Fix: Mutate all (N45Q, N89Q, N138Q, N276Q)
Step 2: Construct optimization
AlphaFold: C-terminus (450-478) disordered
Intracellular loop (340-385) flexible
Fix: Truncate C-terminus, stabilize loop (V351A, L367S, F378A)
Step 3: Preferred orientation strategy
Fix: Use nanodiscs (MSP1D1, POPC:POPG 3:1)
Isotropic shape reduces orientation bias
Step 4: Stability optimization
Orbion predicts agonist stabilizes open state
Fix: Add saturating agonist (100 μM)
Final Construct & Strategy
Residues: 1-449 (truncated)
Mutations: N45Q, N89Q, N138Q, N276Q, V351A, L367S, F378A
Nanodisc reconstitution
Agonist-bound
Results
Expression: 4 mg/L (2× improvement)
Tm: 62°C (+8°C)
Grids: 15 made, first grid showed good particles
Preferred orientation: Solved (angular distribution uniform)
Data: Single session (3 days)
Processing: 800,000 particles
Final: 2.9 Å structure
Cost: $60K, 5 months Outcome: Structure published
Lesson: Computational prediction identified all problems before grid optimization.
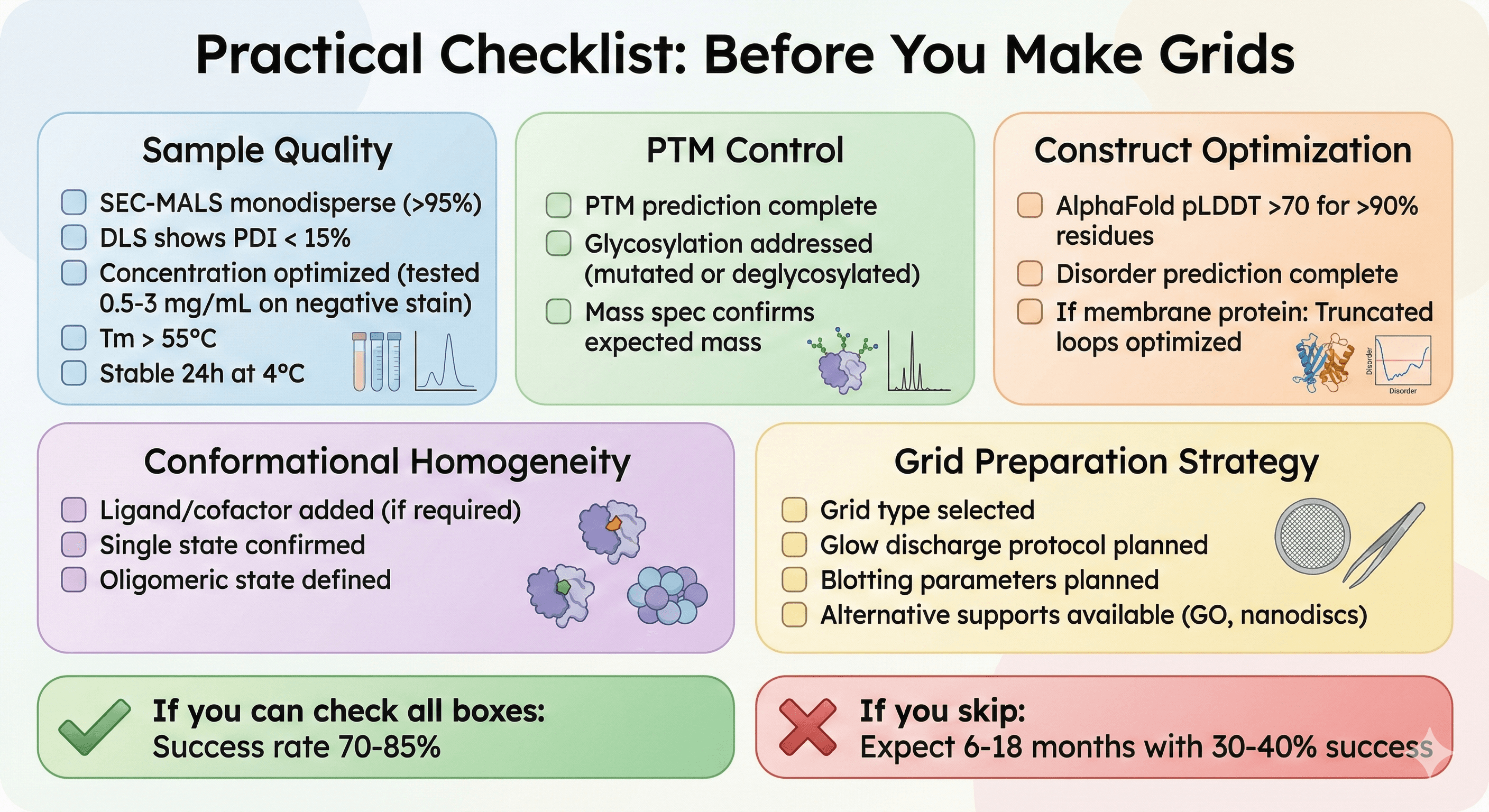
Practical Checklist: Before You Make Grids
☐ Sample Quality
[ ] SEC-MALS monodisperse (>95%)
[ ] DLS shows PDI < 15%
[ ] Concentration optimized (tested 0.5-3 mg/mL on negative stain)
[ ] Tm > 55°C
[ ] Stable 24h at 4°C
☐ PTM Control
[ ] PTM prediction complete
[ ] Glycosylation addressed (mutated or deglycosylated)
[ ] Mass spec confirms expected mass
☐ Construct Optimization
[ ] AlphaFold pLDDT >70 for >90% residues
[ ] Disorder prediction complete
[ ] If membrane protein: Truncated loops optimized
☐ Conformational Homogeneity
[ ] Ligand/cofactor added (if required)
[ ] Single state confirmed
[ ] Oligomeric state defined
☐ Grid Preparation Strategy
[ ] Grid type selected
[ ] Glow discharge protocol planned
[ ] Blotting parameters planned
[ ] Alternative supports available (GO, nanodiscs)
If you can check all boxes: Success rate 70-85%
If you skip: Expect 6-18 months with 30-40% success
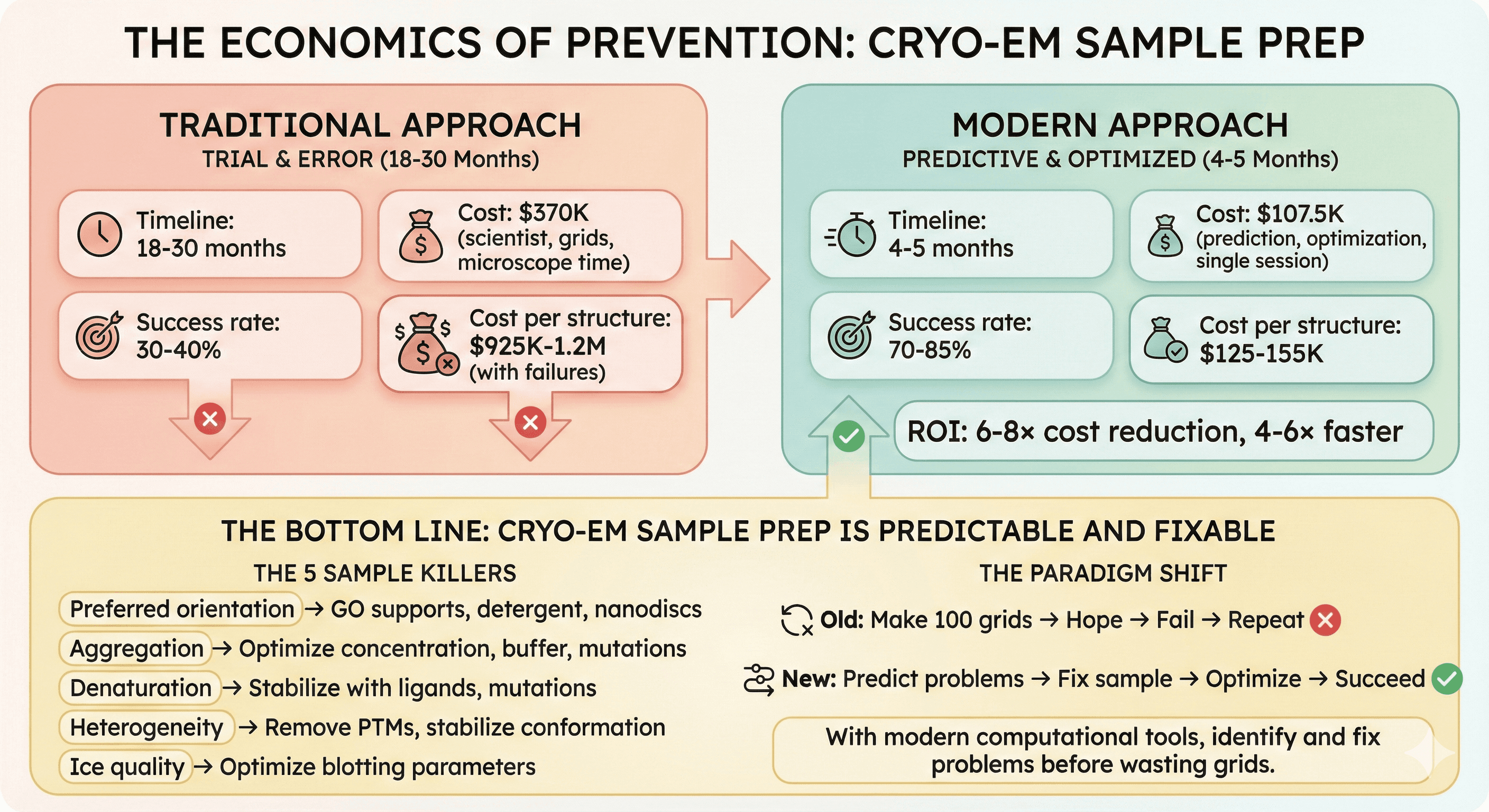
The Economics of Prevention
Traditional Approach
Timeline: 18-30 months Cost: $370K (scientist, grids, microscope time) Success rate: 30-40% Cost per structure: $925K-1.2M (with failures)
Modern Approach
Timeline: 4-5 months Cost: $107.5K (prediction, optimization, single session) Success rate: 70-85% Cost per structure: $125-155K
ROI: 6-8× cost reduction, 4-6× faster
The Bottom Line
Cryo-EM sample preparation is predictable and fixable.
The 5 sample killers:
Preferred orientation → GO supports, detergent, nanodiscs
Aggregation → Optimize concentration, buffer, mutations
Denaturation → Stabilize with ligands, mutations
Heterogeneity → Remove PTMs, stabilize conformation
Ice quality → Optimize blotting parameters
The paradigm shift:
Old: Make 100 grids → Hope → Fail → Repeat
New: Predict problems → Fix sample → Optimize → Succeed
With modern computational tools, identify and fix problems before wasting grids.
Ready to Optimize Your Cryo-EM Sample?
If you're preparing for cryo-EM, Orbion can identify potential problems before you make your first grid.
Orbion provides:
Complete PTM landscape (glycosylation causing heterogeneity)
Construct optimization (disorder, flexible loops)
Aggregation hotspot prediction
Stability optimization (thermostabilizing mutations)
Membrane topology analysis (for nanodiscs)
From sequence to cryo-EM-ready sample in <1 day.

