Blog
The 5 Cryo-EM Sample Killers: Understanding Why Your Particles Won't Align
Jan 5, 2026
You've spent $150K on a high-resolution cryo-EM microscope session. Your protein is pure, stable, and monodisperse. You make grids, collect data, and begin processing. Then you see it: your 2D class averages are a blurry mess. Particles won't align. Half your classes look like noise.
After three days of data collection, you have nothing usable.
Welcome to the most frustrating bottleneck in cryo-EM: sample preparation. While cryo-EM has revolutionized structural biology, the technique's Achilles' heel remains the grid. Get the sample wrong, and $200K of microscope time produces nothing but expensive noise.
This is Part 1 of our cryo-EM troubleshooting guide. Here, we'll diagnose why samples fail. In Part 2, we'll cover how to fix each problem and optimize your workflow.
Key Takeaways
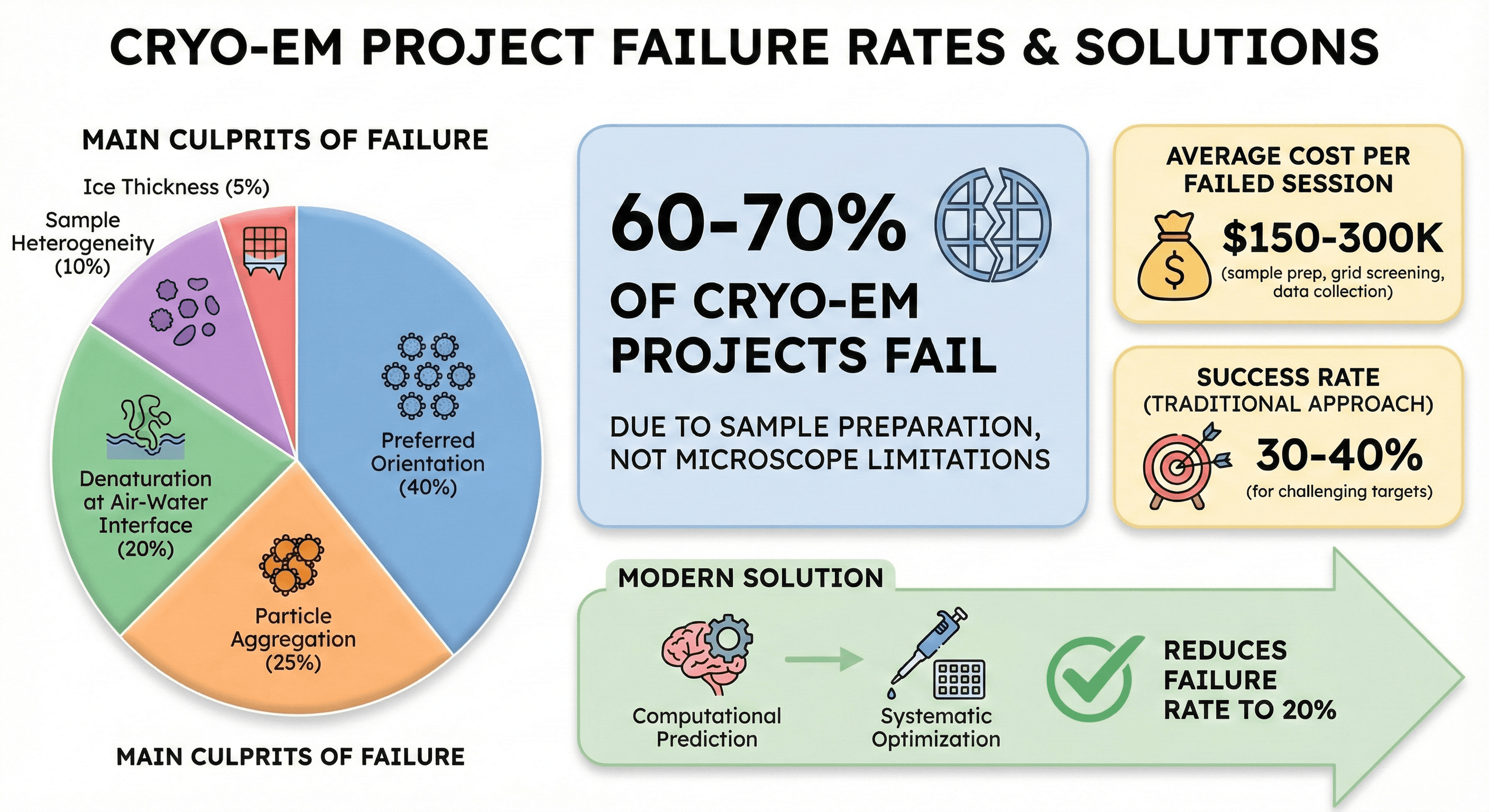
60-70% of cryo-EM projects fail due to sample preparation, not microscope limitations
Main culprits: Preferred orientation (40%), particle aggregation (25%), denaturation at air-water interface (20%), sample heterogeneity (10%), ice thickness (5%)
Average cost per failed session: $150-300K (sample prep, grid screening, data collection)
Success rate: 30-40% for challenging targets with traditional approach
Modern solution: Computational prediction + systematic optimization reduces failure rate to 20%
The Cryo-EM Revolution's Hidden Bottleneck
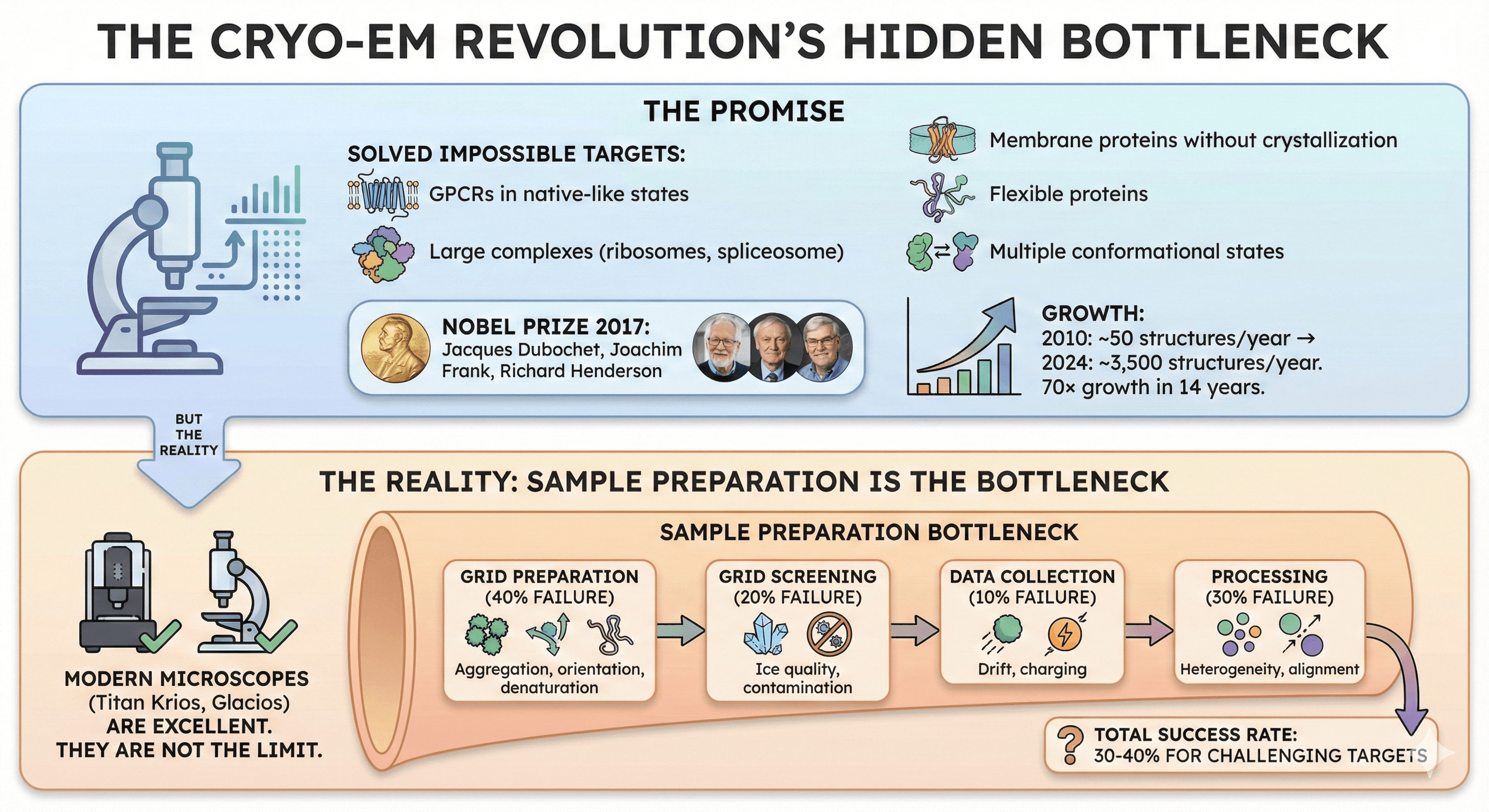
The Promise
Cryo-EM solved the "impossible targets":
GPCRs in native-like states
Large complexes (ribosomes, spliceosome)
Membrane proteins without crystallization
Flexible proteins
Multiple conformational states
Nobel Prize 2017: Jacques Dubochet, Joachim Frank, Richard Henderson
Growth:
2010: ~50 structures/year
2024: ~3,500 structures/year
70× growth in 14 years
The Reality
Cryo-EM success is NOT limited by microscopes. Modern microscopes (Titan Krios, Glacios) are excellent. The bottleneck is sample preparation.
Failure at each stage:
Grid preparation: 40% failure (aggregation, orientation, denaturation)
Grid screening: 20% failure (ice quality, contamination)
Data collection: 10% failure (drift, charging)
Processing: 30% failure (heterogeneity, alignment)
Total success rate: 30-40% for challenging targets
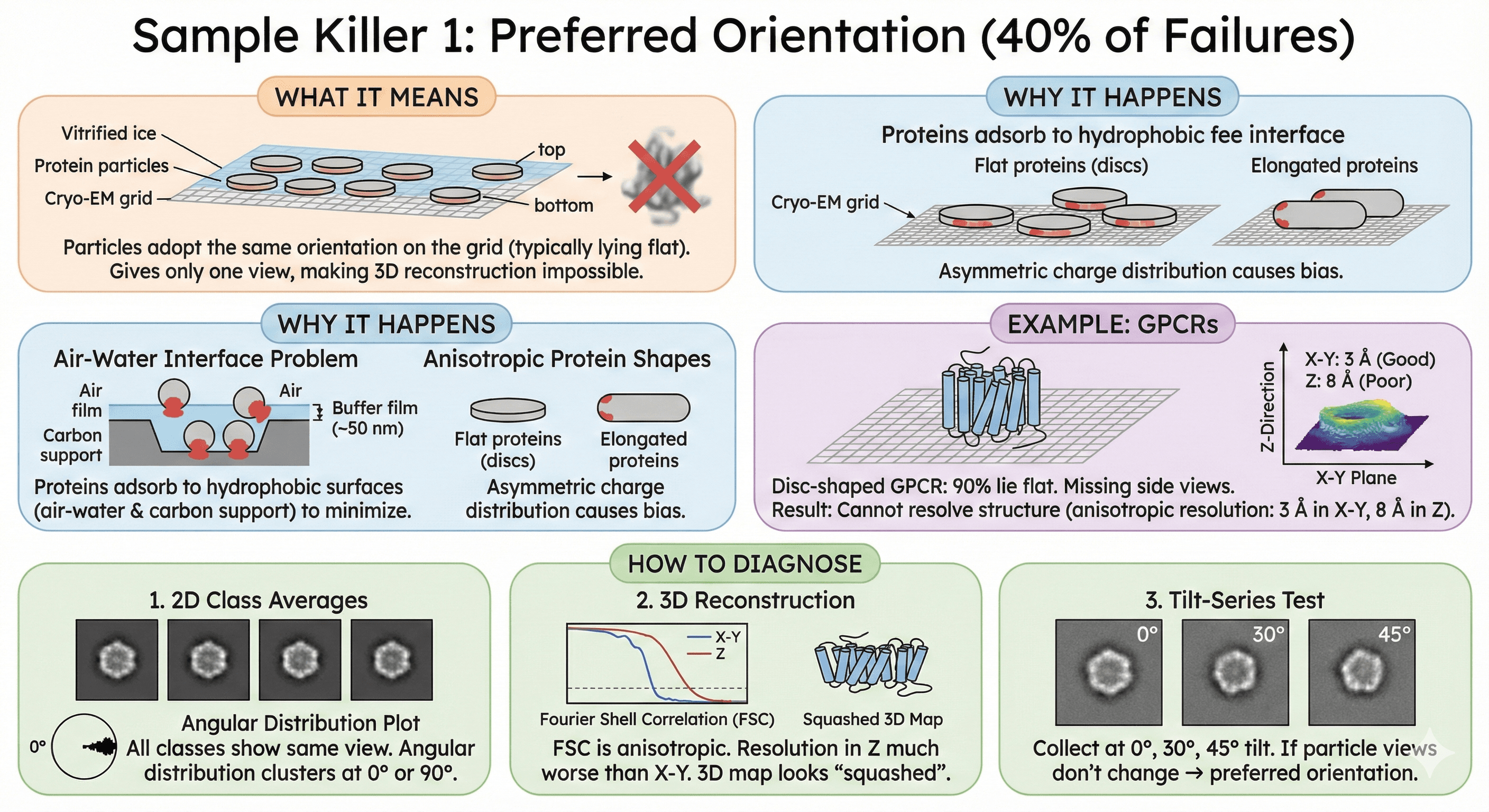
Sample Killer 1: Preferred Orientation (40% of Failures)
What it means: Particles adopt the same orientation on the grid—typically lying flat. This gives you only one view, making 3D reconstruction impossible.
Why It Happens
The air-water interface problem:
When you blot the grid, a thin film of buffer (~50 nm) remains
Creates two hydrophobic surfaces: air-water (top) and carbon support (bottom)
Proteins adsorb to minimize free energy
Result: Proteins orient with most hydrophobic face toward surface
Anisotropic protein shapes:
Flat proteins (discs) lie flat
Elongated proteins align parallel
Asymmetric charge distribution causes bias
Example: GPCRs
Disc-shaped (7-TM cylinder)
90% lie flat on grid
Missing: Side views
Result: Cannot resolve structure (anisotropic resolution: 3 Å in X-Y, 8 Å in Z)
How to Diagnose
1. 2D class averages:
All classes show same view (e.g., all top-down)
No side views, no tilted views
Angular distribution plot clusters at 0° or 90°
2. 3D reconstruction:
Fourier Shell Correlation (FSC) is anisotropic
Resolution in Z-direction much worse than X-Y
3D map looks "squashed"
3. Tilt-series test:
Collect at 0°, 30°, 45° tilt
If particle views don't change → preferred orientation
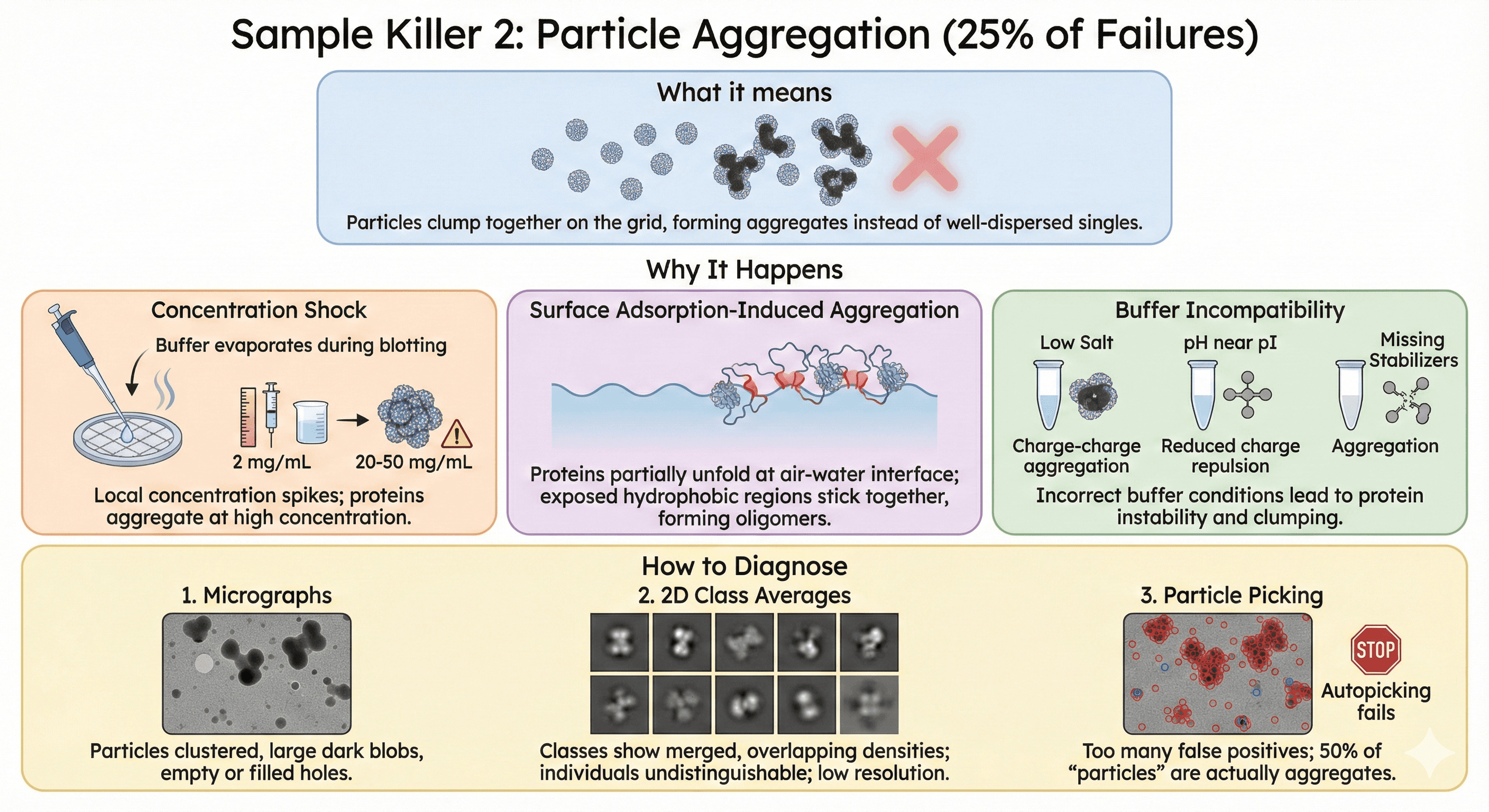
Sample Killer 2: Particle Aggregation (25% of Failures)
What it means: Particles clump together on the grid, forming aggregates instead of well-dispersed singles.
Why It Happens
Concentration shock:
During blotting, buffer evaporates
Local concentration spikes from 2 mg/mL → 20-50 mg/mL
At high concentration, proteins aggregate
Surface adsorption-induced aggregation:
Proteins partially unfold at air-water interface
Exposed hydrophobic regions stick together
Forms oligomers and larger aggregates
Buffer incompatibility:
Low salt → charge-charge aggregation
pH near pI → reduced charge repulsion
Missing stabilizers → aggregation
How to Diagnose
1. Micrographs:
Particles clustered (not evenly distributed)
Large dark blobs instead of individual particles
Holes either empty or completely filled
2. 2D class averages:
Classes show merged particles (overlapping densities)
Cannot distinguish individuals
Low-resolution classes
3. Particle picking:
Autopicking fails (too many false positives)
50% of "particles" are actually aggregates
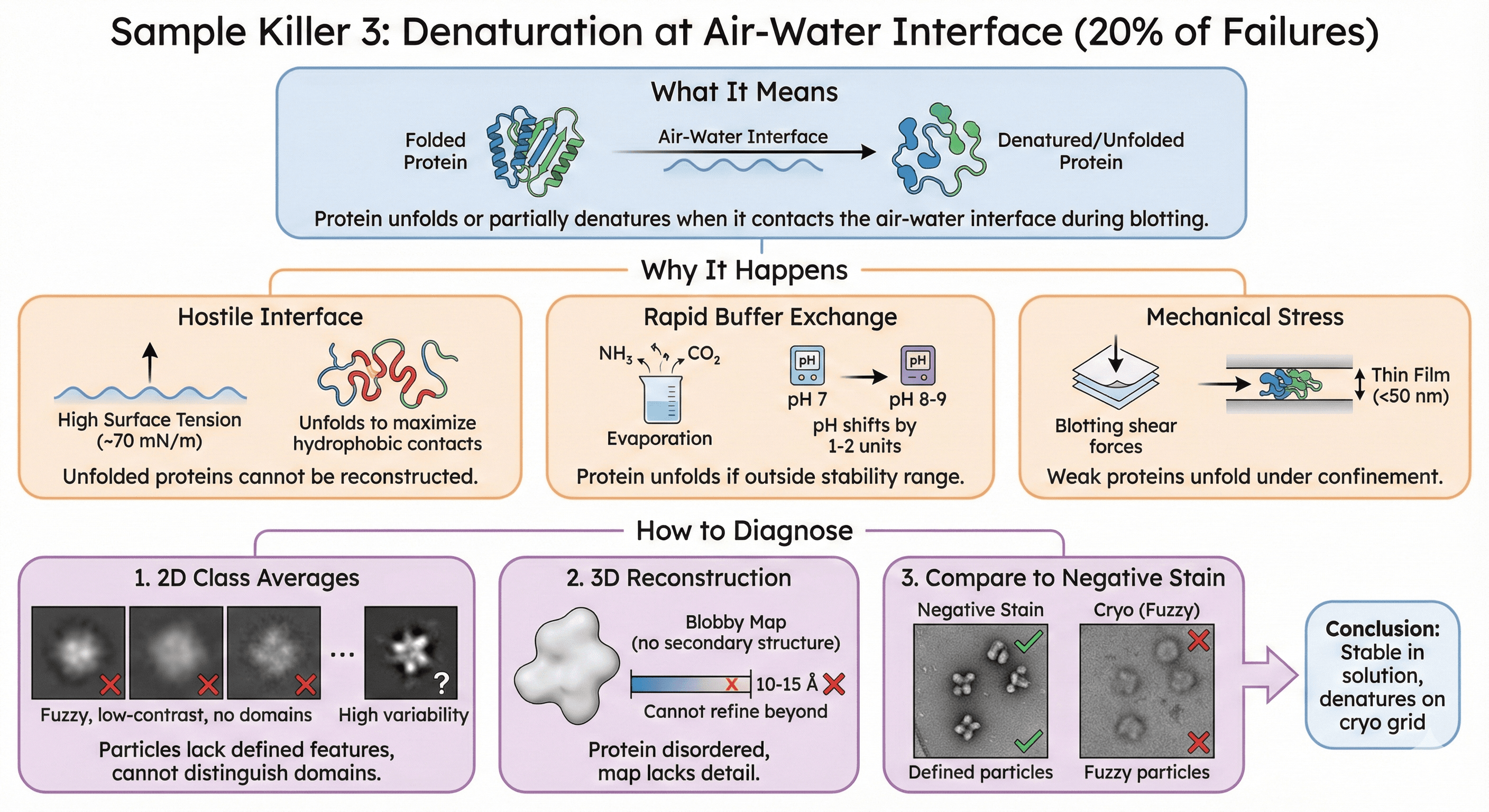
Sample Killer 3: Denaturation at Air-Water Interface (20% of Failures)
What it means: Protein unfolds or partially denatures when it contacts the air-water interface during blotting.
Why It Happens
Hostile interface:
Air-water interface has high surface tension (~70 mN/m)
Proteins unfold to maximize hydrophobic contacts with air
Unfolded proteins cannot be reconstructed
Rapid buffer exchange:
Volatile buffer components evaporate (ammonia, CO₂)
pH shifts by 1-2 units in seconds
Protein unfolds if outside stability range
Mechanical stress:
Blotting applies shear forces
Thin film (<50 nm) confines proteins
Weak proteins unfold
How to Diagnose
1. 2D class averages:
Particles lack defined features (fuzzy, low-contrast)
Cannot distinguish domains
High variability between classes
2. 3D reconstruction:
Cannot refine beyond 10-15 Å (protein disordered)
Map is blobby (no secondary structure)
3. Compare to negative stain:
Negative stain shows defined particles
Cryo shows fuzzy particles
Conclusion: Stable in solution, denatures on cryo grid
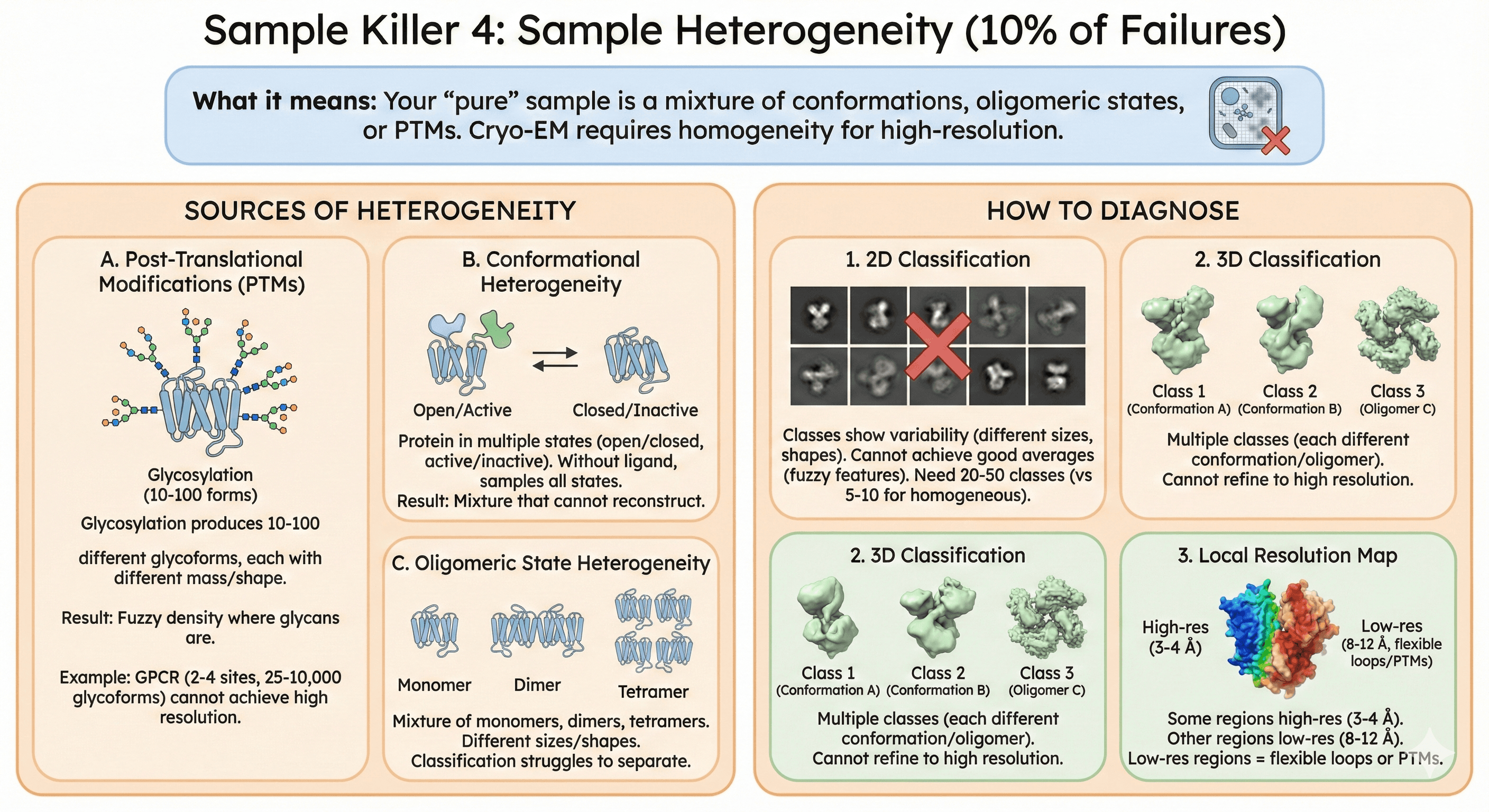
Sample Killer 4: Sample Heterogeneity (10% of Failures)
What it means: Your "pure" sample is a mixture of conformations, oligomeric states, or PTMs. Cryo-EM requires homogeneity for high-resolution.
Sources of Heterogeneity
A. Post-Translational Modifications (PTMs)
The problem:
Glycosylation produces 10-100 different glycoforms
Each has different mass/shape
Result: Fuzzy density where glycans are
Example: GPCR glycosylation
2-4 N-glycosylation sites
Each site: 5-10 glycan structures
Total: 25 to 10,000 glycoforms
Cannot achieve high resolution with this heterogeneity
B. Conformational heterogeneity
Protein in multiple states (open/closed, active/inactive)
Without ligand, samples all states
Result: Mixture that cannot reconstruct
C. Oligomeric state heterogeneity
Mixture of monomers, dimers, tetramers
Different sizes/shapes
Classification struggles to separate
How to Diagnose
1. 2D classification:
Classes show variability (different sizes, shapes)
Cannot achieve good averages (fuzzy features)
Need 20-50 classes (vs 5-10 for homogeneous)
2. 3D classification:
Multiple classes (each different conformation/oligomer)
Cannot refine to high resolution
3. Local resolution map:
Some regions high-res (3-4 Å)
Other regions low-res (8-12 Å)
Low-res regions = flexible loops or PTMs
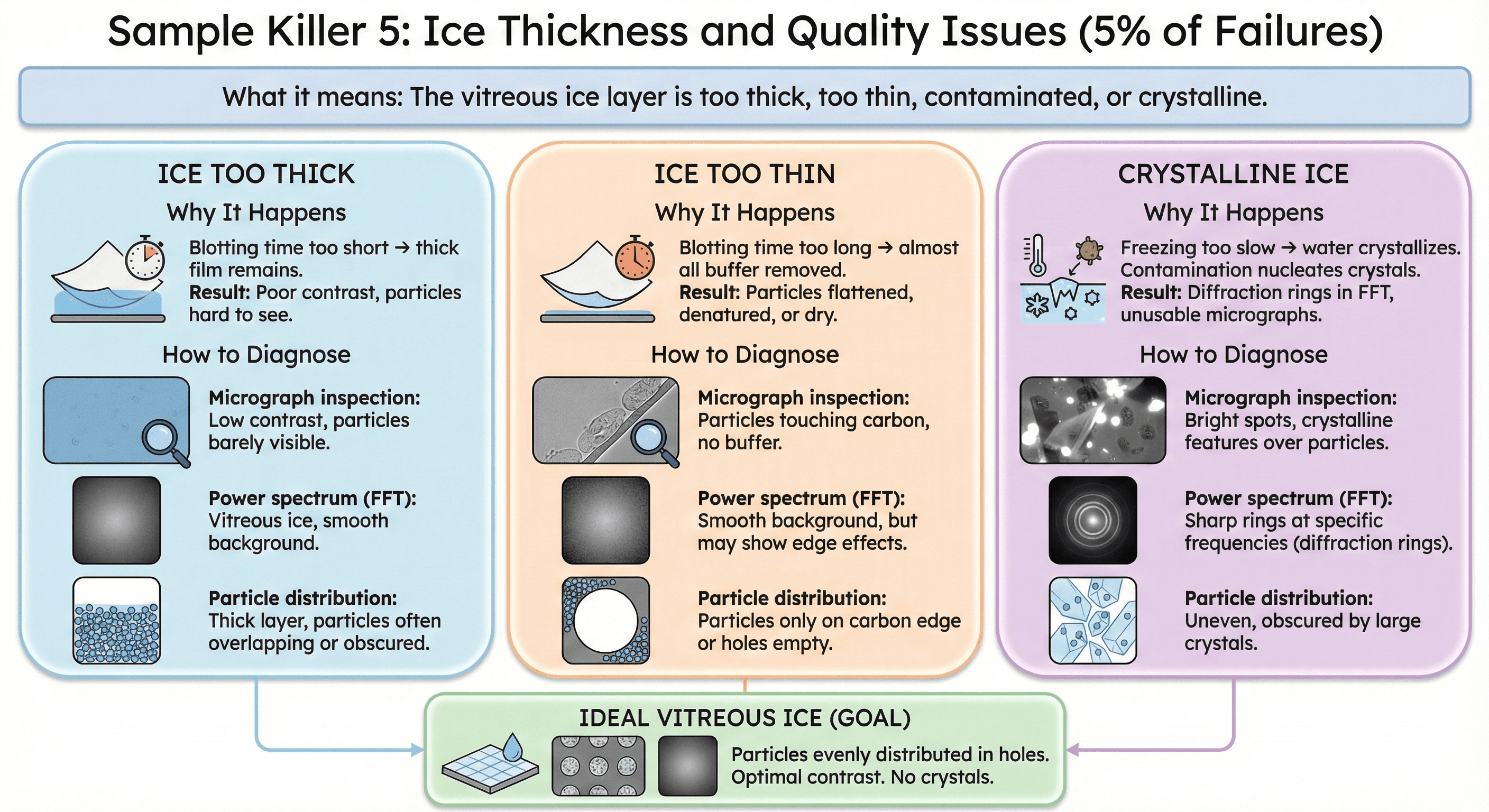
Sample Killer 5: Ice Thickness and Quality Issues (5% of Failures)
What it means: The vitreous ice layer is too thick, too thin, contaminated, or crystalline.
Why It Happens
Ice too thick:
Blotting time too short → thick film remains
Result: Poor contrast, particles hard to see
Ice too thin:
Blotting time too long → almost all buffer removed
Result: Particles flattened, denatured, or dry
Crystalline ice:
Freezing too slow → water crystallizes
Contamination nucleates crystals
Result: Diffraction rings in FFT, unusable micrographs
How to Diagnose
1. Micrograph inspection:
Ice too thick: Low contrast, particles barely visible
Ice too thin: Particles touching carbon, no buffer
Crystalline: Bright spots in FFT, diffraction rings
2. Power spectrum (FFT):
Vitreous ice: Smooth, featureless background
Crystalline ice: Sharp rings at specific frequencies
3. Particle distribution:
Good ice: Particles evenly distributed in holes
Bad ice: Particles only on carbon edge or holes empty
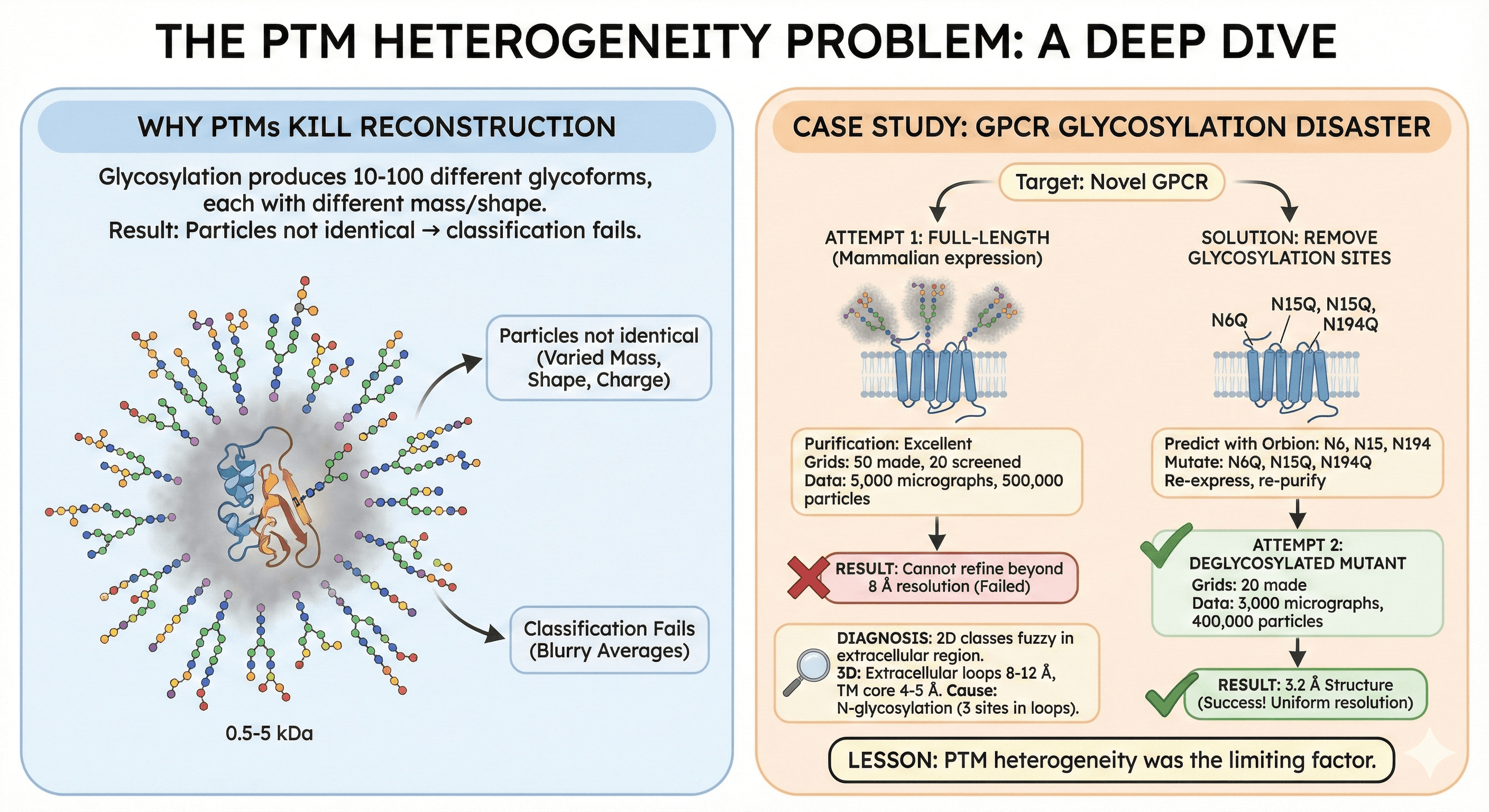
The PTM Heterogeneity Problem: A Deep Dive
This is where most projects underestimate the challenge.
Why PTMs Kill Reconstruction
Glycosylation:
Single site can produce 10-50 glycoforms
Each has different mass (0.5-5 kDa)
Each has different shape and charge
Result: Particles not identical → classification fails
Case Study: GPCR Glycosylation Disaster
Target: Novel GPCR
Attempt 1: Full-length (mammalian expression)
Purification: Excellent
Grids: 50 made, 20 screened
Data: 5,000 micrographs, 500,000 particles
Result: Cannot refine beyond 8 Å resolution
Diagnosis:
2D classes fuzzy in extracellular region
3D: Extracellular loops 8-12 Å, TM core 4-5 Å
Cause: N-glycosylation (3 sites in loops)
Solution: Remove glycosylation sites
Predict with Orbion: N6, N15, N194
Mutate: N6Q, N15Q, N194Q
Re-express, re-purify
Attempt 2: Deglycosylated mutant
Grids: 20 made
Data: 3,000 micrographs, 400,000 particles
Result: 3.2 Å structure (uniform resolution)
Lesson: PTM heterogeneity was the limiting factor.
Understanding Your Problem: Quick Diagnostic
Symptom | Likely Problem | Quick Test |
|---|---|---|
All 2D classes show same view | Preferred orientation | Check angular distribution |
Particles clustered in micrographs | Aggregation | DLS at crystallization concentration |
2D classes fuzzy, no features | Denaturation | Compare to negative stain |
Many 3D classes needed | Heterogeneity | Check for PTMs, oligomers |
Bright spots in FFT | Crystalline ice | Check blotting parameters |
Very dark or very light holes | Ice thickness | Optimize blot time |
Key Takeaway
Cryo-EM sample preparation isn't random trial-and-error. It's a predictable engineering problem with known failure modes:
Preferred orientation (40%): Particles lie flat
Aggregation (25%): Clumping at high concentration
Denaturation (20%): Unfolding at air-water interface
Heterogeneity (10%): PTMs, conformational states
Ice quality (5%): Too thick, thin, or crystalline
Understanding your failure mode is the first step to solving it.

