Blog
Finding Druggable Pockets When the Active Site Is Too Conserved
Jan 26, 2026
Your kinase inhibitor is potent. Sub-nanomolar IC50. Beautiful binding to the ATP pocket. There's just one problem: it also inhibits 47 other kinases in the same family. The selectivity panel is a sea of red. Your drug candidate is toxic before it reaches the clinic.
The ATP binding site was the obvious target. It's also the most conserved pocket across the kinome. Every kinase binds ATP the same way. Your molecule can't tell them apart.
This is the selectivity problem—and it's why drug discovery increasingly looks beyond the active site.
Key Takeaways
Active sites are often too conserved for selective drug design (90%+ sequence identity across families)
Allosteric sites offer selectivity because they're under less evolutionary pressure
Cryptic pockets exist in many proteins but only appear in certain conformations
Protein-protein interaction sites are emerging targets but require different approaches
Computational methods can identify non-obvious binding sites before you screen
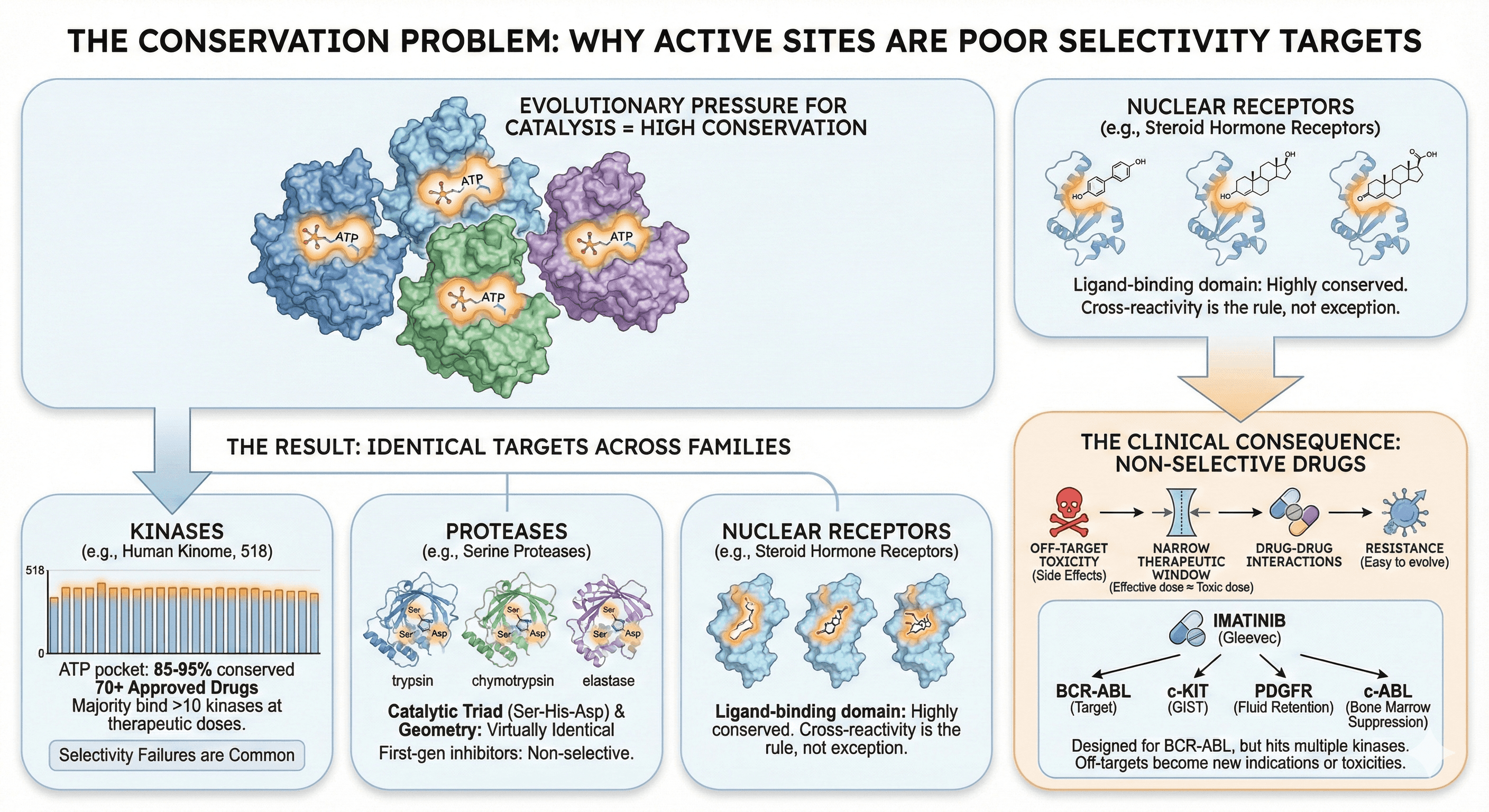
The Conservation Problem
Why Active Sites Are Poor Selectivity Targets
Evolution has shaped active sites for one purpose: catalysis. The residues that bind substrates and transition states are under intense selection pressure. Change them, and the enzyme dies.
The result:
Active sites are the most conserved regions of protein families
ATP-binding pockets across 500+ human kinases share >80% sequence identity
Substrate-binding sites in protease families are nearly identical
The pocket you're targeting looks the same in your target and its closest 50 relatives
The Numbers
Human kinome (Manning et al., 2002):
518 protein kinases
ATP pocket: 85-95% conserved across families
Approved kinase inhibitors: 70+ drugs
Selectivity failures: The majority bind >10 kinases at therapeutic doses (Klaeger et al., 2017)
Proteases:
Serine proteases share the catalytic triad (Ser-His-Asp)
Active site geometry: virtually identical
First-generation inhibitors: almost universally non-selective
Nuclear receptors:
Ligand-binding domain: highly conserved
Steroid hormone receptors: cross-reactivity is the rule, not exception
The Clinical Consequence
Non-selective drugs cause:
Off-target toxicity: Hitting related proteins causes side effects
Narrow therapeutic window: The dose that's effective is close to toxic
Drug-drug interactions: Multiple kinases affected by the same drug
Resistance: Easy to evolve resistance when many targets exist
Example: First-generation tyrosine kinase inhibitors
Imatinib (Gleevec) was designed against BCR-ABL kinase (Druker et al., 2001). But it also hits:
c-KIT (useful for GIST tumors)
PDGFR (causes fluid retention)
c-ABL in normal cells (bone marrow suppression)
The "off-target" effects became indications (GIST) or dose-limiting toxicities (edema, cytopenias).
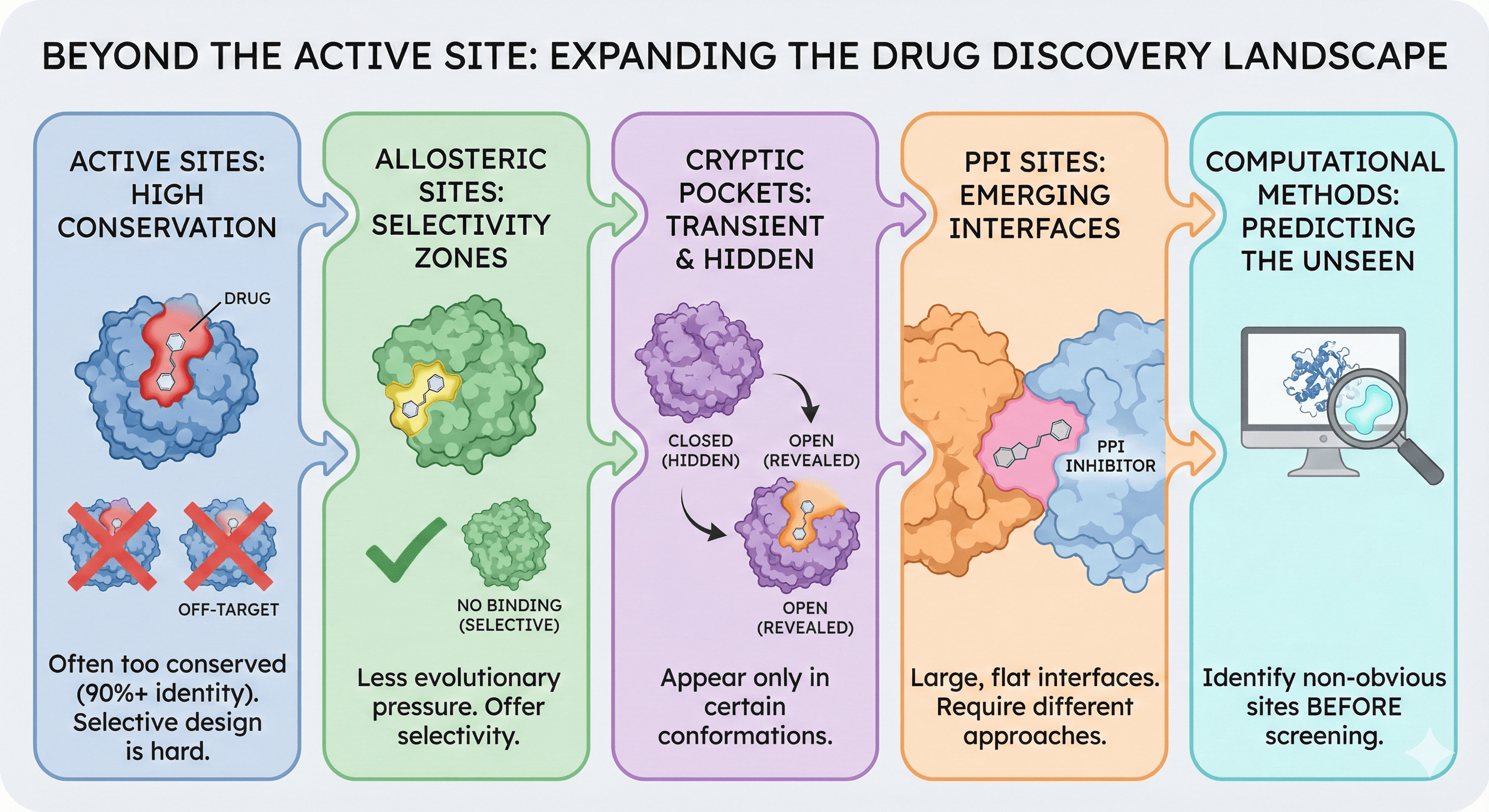
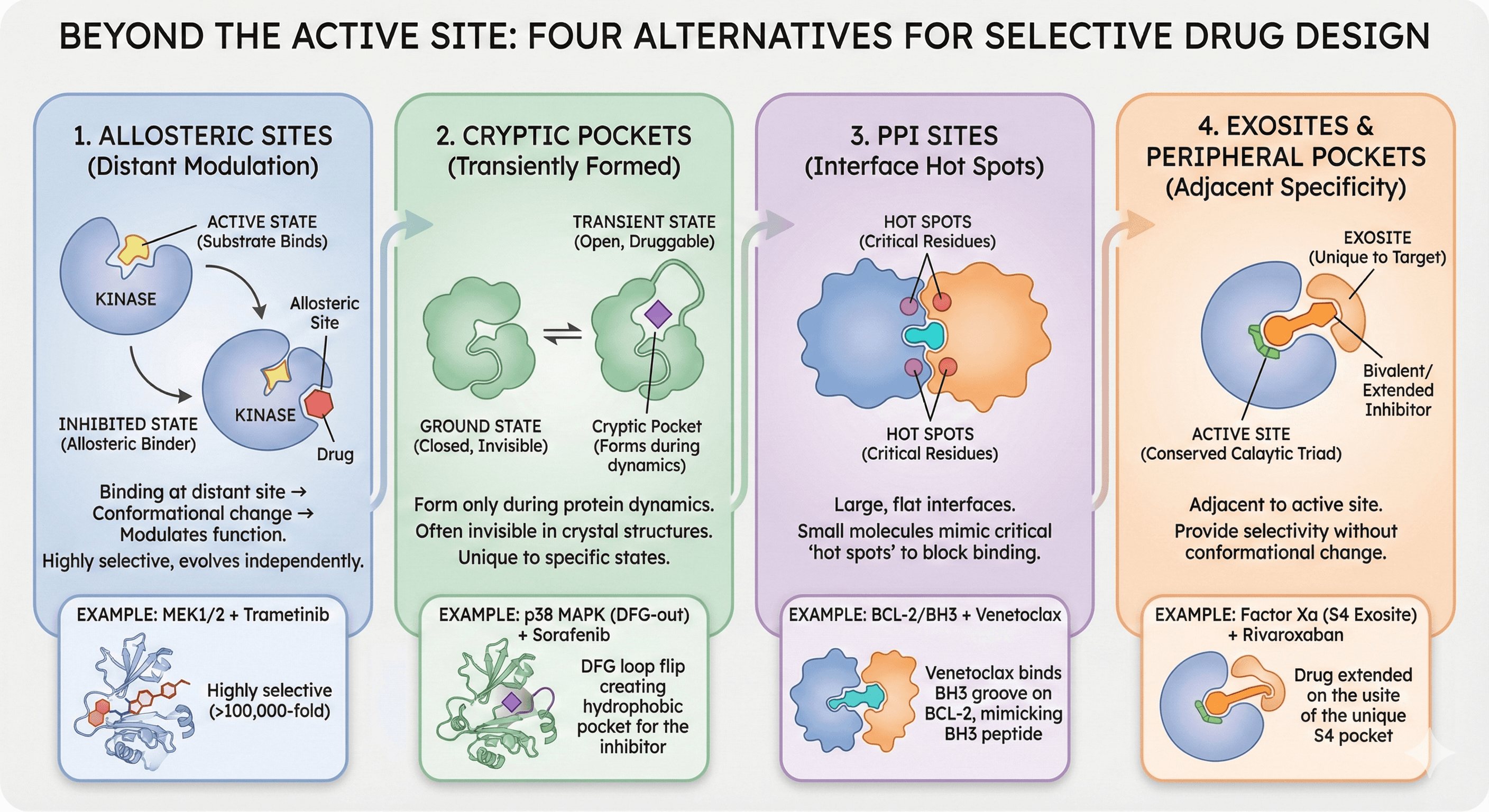
Beyond the Active Site: Four Alternatives
Alternative 1: Allosteric Sites
What they are: Binding sites distant from the active site that modulate function through conformational change.
Why they're selective:
Not under substrate-binding pressure
Evolve independently of catalytic function
Often unique to specific proteins or subfamilies
How allosteric drugs work:
Small molecule binds to allosteric site
Binding causes conformational change
Conformational change alters active site (activates or inhibits)
Function is modulated without competing with substrate
Examples of allosteric successes:
Drug | Target | Allosteric Site | Advantage |
|---|---|---|---|
Trametinib | MEK1/2 | Allosteric pocket | Highly selective for MEK |
Asciminib | BCR-ABL | Myristate pocket | Overcomes imatinib resistance |
Venetoclax | BCL-2 | BH3 groove | First-in-class protein-protein disruptor |
The selectivity advantage:
Trametinib IC50 for MEK1: 0.7 nM
Next closest kinase: >100,000-fold selectivity
Compare to ATP-competitive inhibitors: often <10-fold selectivity
Alternative 2: Cryptic Pockets
What they are: Binding sites that don't exist in the ground-state structure but form transiently during protein dynamics.
Why they're valuable:
Not visible in crystal structures (which capture one conformation)
Unique to specific conformational states
Often druggable despite appearing "absent"
How cryptic pockets form:
Side chain movements expose buried cavities
Domain motions open transient channels
Loop movements create temporary binding sites
Ligand binding induces pocket formation (induced fit)
Detection methods:
Molecular dynamics: Simulate protein motion, analyze transient cavities
MSA subsampling: Alternative AlphaFold conformations
Ensemble docking: Screen against multiple structures
Fragment screening: Small molecules can "trap" transient states
Example: p38 MAPK
The DFG-out conformation creates an allosteric pocket:
DFG-in (active): No allosteric pocket visible
DFG-out (inactive): Large hydrophobic pocket opens
Type II inhibitors (e.g., sorafenib) bind this cryptic pocket
Much greater selectivity than Type I (ATP-competitive) inhibitors
Alternative 3: Protein-Protein Interaction (PPI) Sites
What they are: Interfaces where proteins contact each other, now targetable by small molecules.
Why they were considered "undruggable":
Large, flat interfaces (1000-2000 Ų)
No obvious pocket
Hot spots distributed across interface
Why that changed:
Hot spots (critical residues) contribute most binding energy
Small molecules can mimic hot spot interactions
Fragment-based approaches can find starting points
Macrocycles and peptide mimetics span larger areas
Success stories:
Drug | PPI Target | Mechanism |
|---|---|---|
Venetoclax | BCL-2/BH3 | Mimics BH3 peptide binding |
Nutlins | MDM2/p53 | Blocks p53 binding groove |
BET inhibitors | BRD4/acetyl-lysine | Displaces histone binding |
The hot spot principle:
Only 5-10% of interface residues contribute >75% of binding energy. These "hot spots" are:
Often hydrophobic (Trp, Tyr, Phe)
Clustered spatially
Potentially druggable with small molecules
Alternative 4: Exosites and Peripheral Pockets
What they are: Binding sites adjacent to (but distinct from) the active site.
How they differ from allosteric sites:
Physically closer to active site
May partially overlap with substrate binding
Can provide selectivity without requiring conformational change
Example: Factor Xa exosite
Factor Xa has:
Active site: Serine protease catalytic triad (conserved across coagulation cascade)
S4 exosite: Adjacent pocket unique to Factor Xa
Drug strategy:
First-generation: Active site inhibitors (non-selective, bleeding risk)
Rivaroxaban: Extends into S4 pocket (selective for Factor Xa, safer)
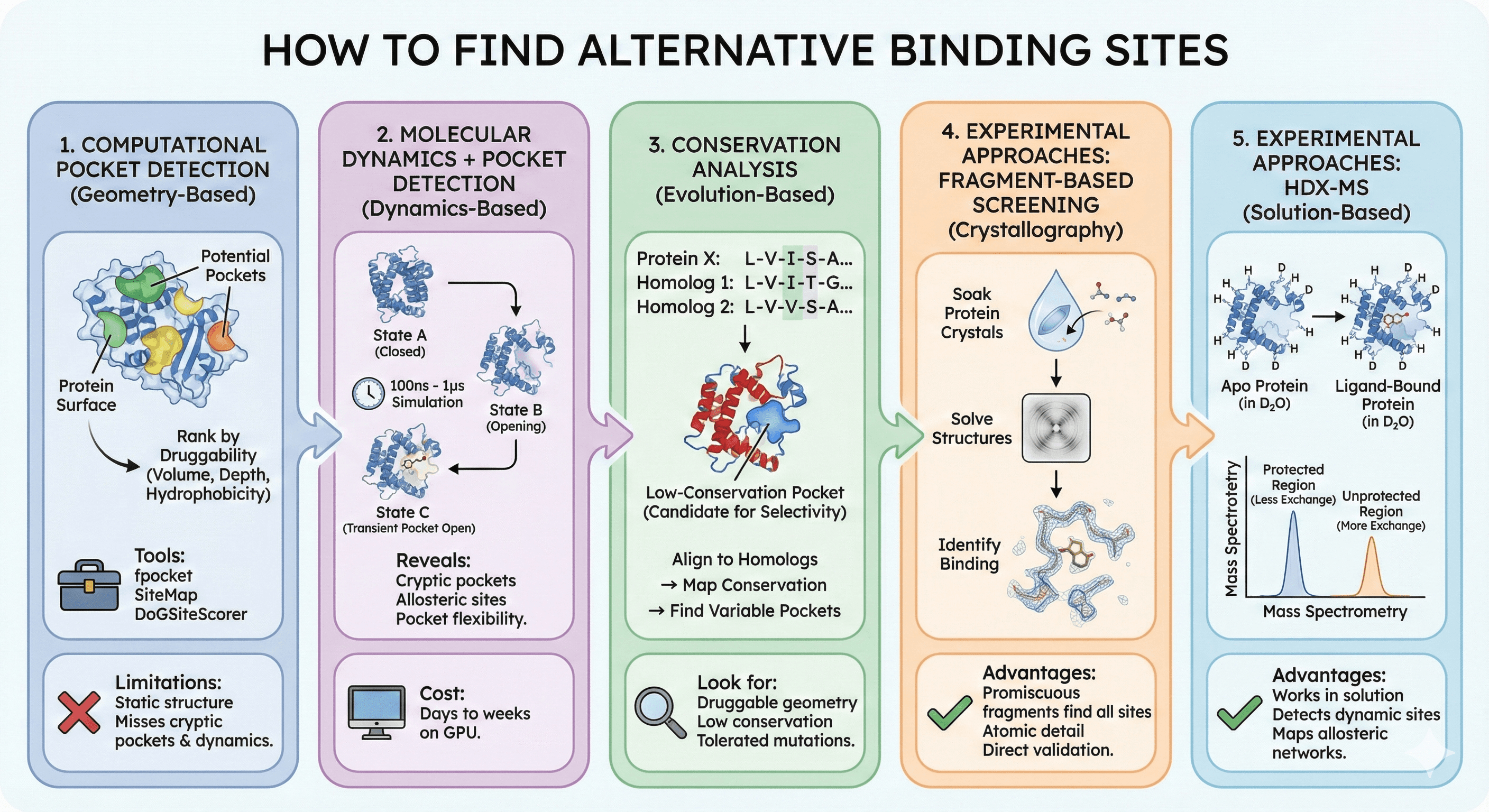
How to Find Alternative Binding Sites
Computational Pocket Detection
Geometry-based methods:
Scan protein surface for concave regions
Calculate pocket volume and depth
Rank by "druggability" (hydrophobicity, enclosure)
Tools:
fpocket: Fast pocket detection from structure
SiteMap: Commercial, predicts druggability
DoGSiteScorer: Machine learning-enhanced detection
Limitations:
Only detect pockets visible in the input structure
Miss cryptic pockets and conformational changes
Don't account for pocket dynamics
Molecular Dynamics + Pocket Detection
The approach:
Run MD simulation (100 ns - 1 μs)
Cluster conformations
Detect pockets in each cluster
Find pockets that appear transiently
What this reveals:
Cryptic pockets that open during dynamics
Allosteric sites that breathe
Flexibility of known pockets
Druggability of transient states
Computational cost: Days to weeks on GPU clusters
Conservation Analysis
The principle: Allosteric sites are less conserved than active sites.
The workflow:
Align protein to homologs
Map conservation onto structure
Find pockets in low-conservation regions
These are candidates for selective drug design
Tools:
ConSurf: Conservation mapping onto structure
BLAST + structure alignment: Manual analysis
Conservation scores from MSAs
What to look for:
Druggable pocket (geometry, hydrophobicity)
Low conservation in that pocket
Not required for folding (mutations tolerated)
Experimental Approaches
Fragment-based screening:
Soak protein crystals with fragment libraries (small, diverse molecules)
Solve structures
Find where fragments bind
Many fragments find allosteric sites and cryptic pockets
Advantages:
Fragments are promiscuous—they find all binding sites
X-ray crystallography provides atomic detail
Experimental validation of binding
Hydrogen-deuterium exchange (HDX-MS):
Expose protein to D2O with and without ligand
Measure which regions are protected from exchange
Protected regions indicate binding site
Advantages:
Works in solution (no crystallization)
Detects dynamic binding sites
Can map allosteric networks
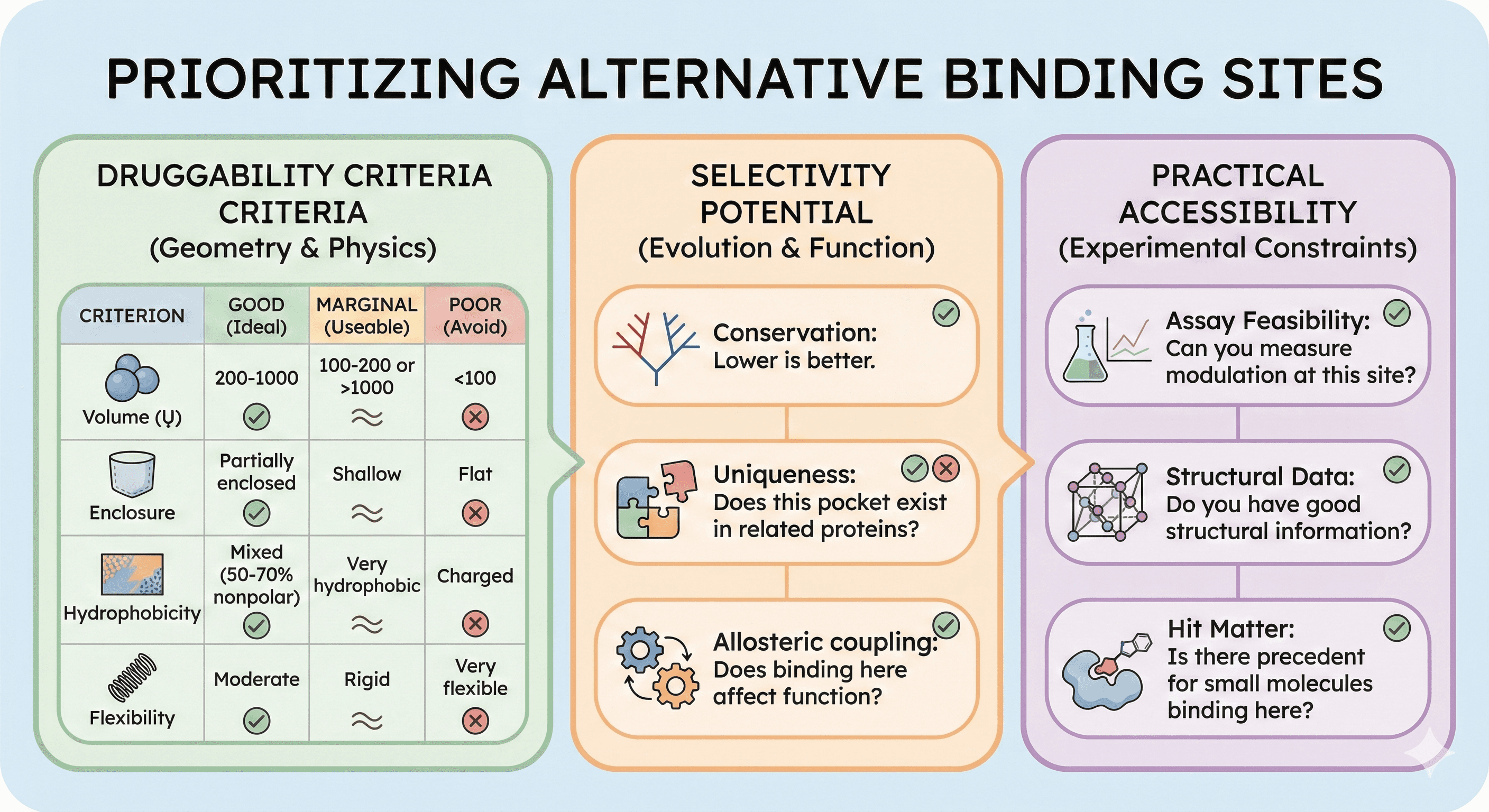
Prioritizing Alternative Sites
Not all pockets are druggable. Use these criteria to prioritize:
Druggability Criteria
Criterion | Good | Marginal | Poor |
|---|---|---|---|
Volume | 200-1000 ų | 100-200 Ų or >1000 ų | <100 ų |
Enclosure | Partially enclosed | Shallow | Flat |
Hydrophobicity | Mixed (50-70% nonpolar) | Very hydrophobic | Charged |
Flexibility | Moderate | Rigid | Very flexible |
Selectivity Potential
Score each pocket for selectivity advantage:
Conservation: Lower is better
Uniqueness: Does this pocket exist in related proteins?
Allosteric coupling: Does binding here affect function?
Practical Accessibility
Consider experimental constraints:
Assay feasibility: Can you measure modulation at this site?
Structural data: Do you have good structural information?
Hit matter: Is there precedent for small molecules binding here?
Case Study: From ATP Site Failure to Allosteric Success
The Challenge
Target: A kinase upregulated in cancer.
First attempt: ATP-competitive inhibitor
IC50: 5 nM against target
But: IC50 <100 nM against 23 related kinases
Clinical result: Dose-limiting toxicity from off-target effects
The Alternative Site Search
Step 1: Conservation analysis
Aligned target to 50 closest kinases
Mapped conservation onto structure
ATP pocket: 92% conserved (expected)
Helix αC region: 40% conserved (interesting)
Myristate pocket: 25% conserved (very interesting)
Step 2: Pocket detection
Ran fpocket on crystal structure
Found 6 putative binding sites
Site 3 overlapped with low-conservation region
Site 5 was a cryptic pocket (only in one crystal form)
Step 3: MD simulation
Ran 500 ns simulation
Site 5 opened transiently (30% of frames)
When open, it connected to the ATP site
Allosteric coupling predicted
The Solution
Strategy: Design molecule that binds Site 5 (cryptic pocket) and stabilizes inactive conformation.
Result:
New compound: IC50 12 nM for target
Selectivity: >1000-fold over all related kinases
Mechanism: Stabilizes inactive conformation, blocks activation
Clinical outcome:
Tolerated at therapeutic doses
No dose-limiting kinase-related toxicity
Proceeded to Phase II
The Lesson
The ATP pocket was a trap. It was obvious, druggable, and led to a potent compound—that was useless in humans. The real opportunity was hidden in a less conserved region that required computational analysis to identify.
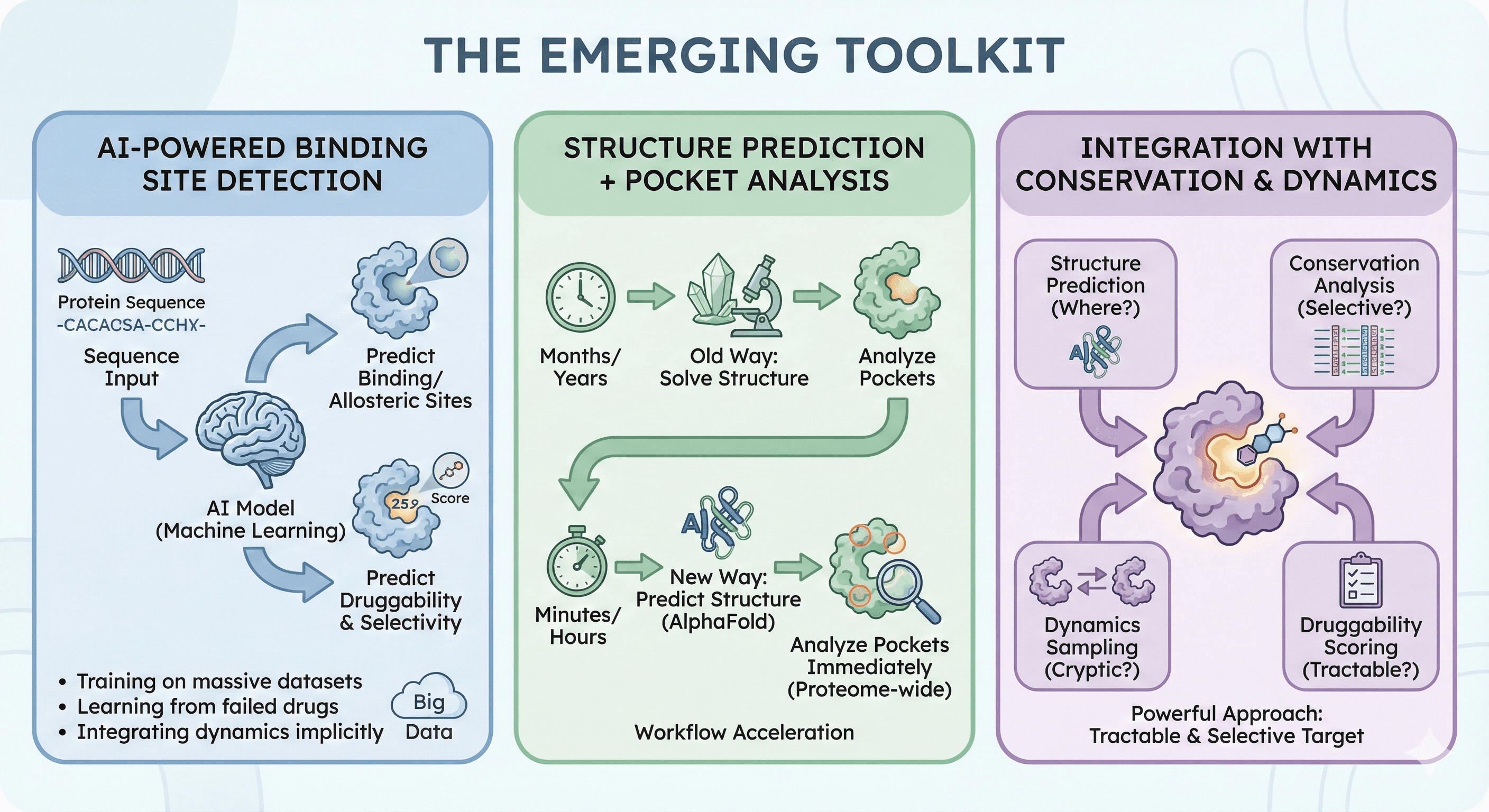
The Emerging Toolkit
AI-Powered Binding Site Detection
Modern machine learning approaches can:
Predict binding sites from sequence (no structure needed)
Identify likely allosteric sites
Predict druggability from learned features
Suggest modifications for selectivity
What's changing:
Training on massive datasets of protein-ligand interactions
Learning from failed drugs (what not to target)
Integrating dynamics implicitly through training data
Structure Prediction + Pocket Analysis
With AlphaFold:
Generate structure in seconds
Analyze for binding sites immediately
Run on the proteome, not just solved structures
The workflow acceleration:
Old way: Solve structure (months/years) → analyze pockets
New way: Predict structure (minutes) → analyze pockets (hours)
Integration with Conservation and Dynamics
The most powerful approach combines:
Structure prediction: Where are the pockets?
Conservation analysis: Which pockets are selective?
Dynamics sampling: What cryptic pockets exist?
Druggability scoring: Which pockets are tractable?
Practical Workflow: Finding Your Alternative Site
Phase 1: Initial Pocket Survey (Day 1)
Get or predict structure
PDB structure if available
AlphaFold prediction if not
Run pocket detection
fpocket or equivalent
List all detected pockets
Map conservation
Align to homologs
Calculate per-residue conservation
Overlay on pockets
Preliminary ranking
Active site: Known, but likely non-selective
Alternative pockets: Ranked by druggability + low conservation
Phase 2: Deeper Analysis (Week 1)
Dynamics (if resources allow)
MD simulation (100 ns minimum)
Cluster conformations
Detect cryptic pockets
Allosteric network analysis
Which residues couple to active site?
Do any alternative pockets include coupled residues?
Literature check
Any known allosteric modulators?
Fragment screening data available?
Mutations in this region affect function?
Phase 3: Experimental Validation (Month 1)
Fragment screening
If you can produce protein: X-ray crystallography with fragments
If not: SPR or DSF with fragment libraries
Functional assay
Can you detect allosteric modulation?
What's the direction (inhibition, activation)?
Hit validation
Confirm binding at alternative site
Measure selectivity improvement
The Bottom Line
Active sites are the obvious targets—and that's the problem. Evolution has optimized them for function, making them nearly identical across protein families. Drugs targeting active sites are potent but non-selective.
The alternative sites (allosteric pockets, cryptic cavities, PPI interfaces) are less obvious but offer the selectivity that active sites can't provide:
Site Type | Conservation | Selectivity Potential | Druggability |
|---|---|---|---|
Active site | Very high | Low | High |
Allosteric site | Moderate | High | Moderate |
Cryptic pocket | Low | Very high | Variable |
PPI interface | Low | Very high | Challenging |
The drug discovery field is shifting from "find the active site, design an inhibitor" to "find the selective site, even if it's not obvious."
Computational Binding Site Identification
For researchers looking to explore alternative binding sites, platforms like Orbion can identify binding sites beyond the obvious active site location. The platform's binding site prediction:
Detects multiple putative binding sites across the protein surface
Maps predictions to the 3D structure for visualization
Provides a starting point for allosteric and alternative site exploration
Combined with conservation analysis and experimental validation, this enables a rational workflow for escaping the selectivity trap that active site targeting inevitably creates.
References
Manning G, et al. (2002). The protein kinase complement of the human genome. Science, 298(5600):1912-1934. Link
Klaeger S, et al. (2017). The target landscape of clinical kinase drugs. Science, 358(6367):eaan4368. Link
Druker BJ, et al. (2001). Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. New England Journal of Medicine, 344(14):1031-1037. Link
Wu P, et al. (2015). Allosteric small-molecule kinase inhibitors. Pharmacology & Therapeutics, 156:59-68. Link
Nussinov R & Tsai CJ. (2013). Allostery in disease and in drug discovery. Cell, 153(2):293-305. Link
Le Guilloux V, et al. (2009). Fpocket: an open source platform for ligand pocket detection. BMC Bioinformatics, 10:168. Link
Fang Z, et al. (2015). Strategies for the selective regulation of kinases with allosteric modulators. Nature Chemical Biology, 11:452-460. Link