Blog
The Hidden Link Between Disorder, Aggregation, and Failed Purifications
Jan 23, 2026
Your protein looked perfect after affinity chromatography. Eluted as a single peak. Then you concentrated it for crystallization trials, walked away for lunch, and came back to a cloudy tube. By the time you ran the gel filtration, half your protein had vanished into the void volume—aggregated, irreversibly.
The frustrating part: this was predictable. The aggregation wasn't random. It was written into the sequence, hidden in the interplay between three properties that most researchers analyze separately but are fundamentally connected: disorder, aggregation propensity, and membrane topology.
Key Takeaways
Disorder exposes aggregation-prone regions that are normally buried in folded proteins
Disordered termini are the #1 cause of post-purification aggregation
Membrane proteins misbehave because their hydrophobic TM helices are aggregation hotspots outside the bilayer
The same hydrophobic patches that drive aggregation also drive membrane insertion—they're two sides of the same coin
Unified analysis of all three properties reveals rescue strategies that single-property predictions miss
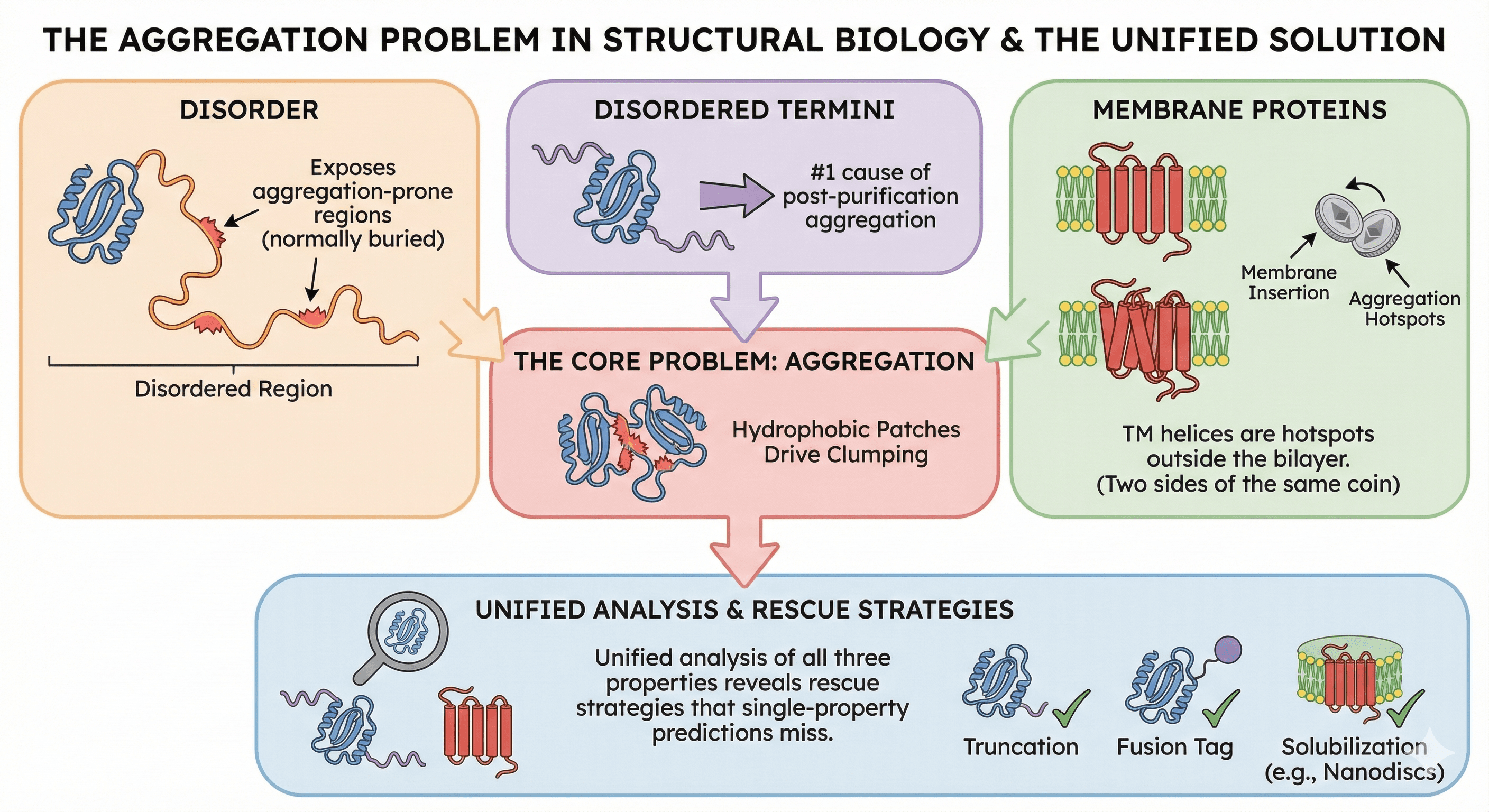
The Hidden Connection
Most prediction tools treat disorder, aggregation, and membrane topology as separate problems:
Use IUPred for disorder
Use AGGRESCAN for aggregation
Use TMHMM for topology
But in the biophysics of your protein, these aren't separate. They're deeply connected through one principle: hydrophobicity and its exposure to water.
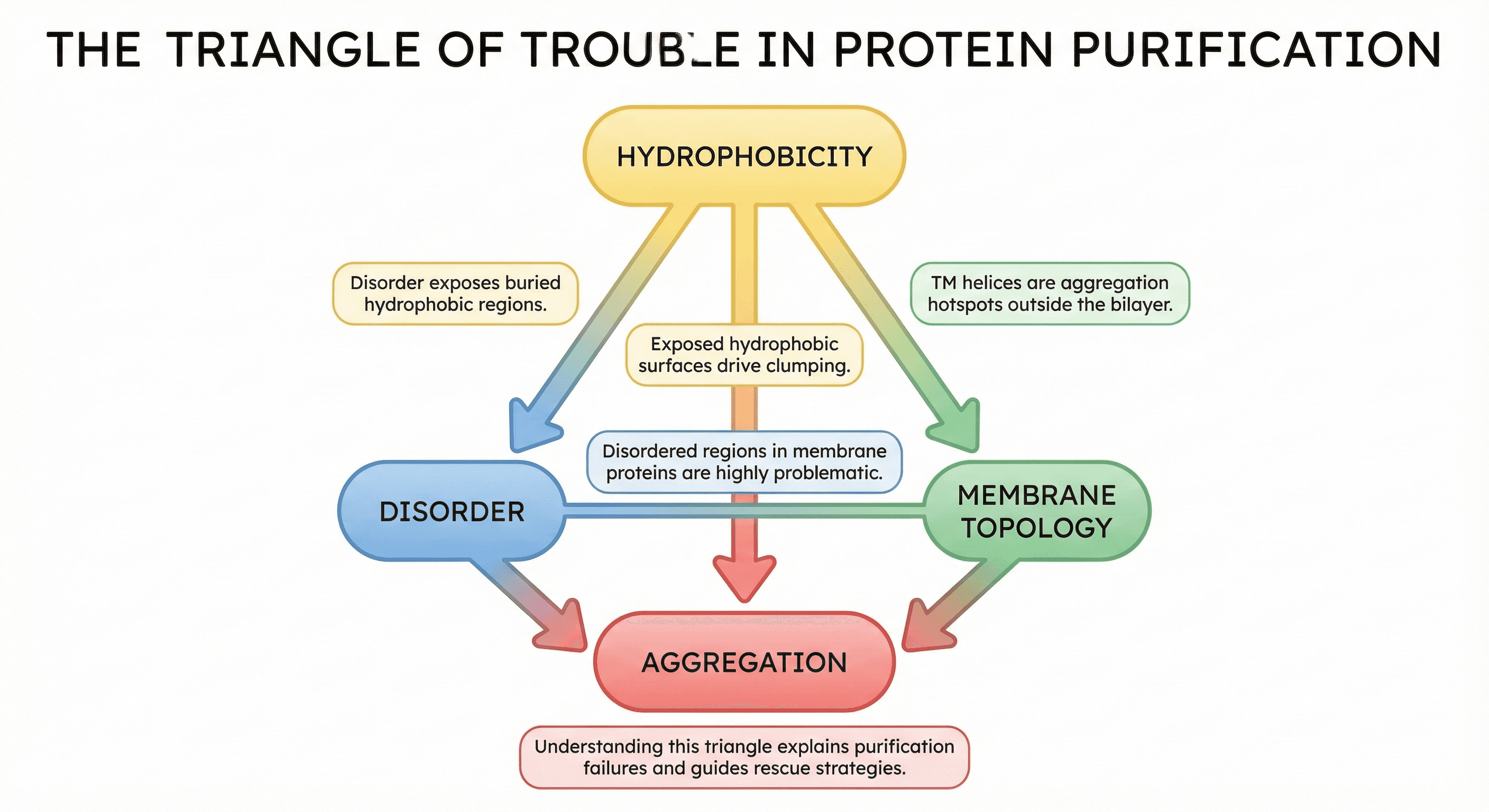
The Triangle of Trouble
The connections:
Disorder exposes hydrophobic regions → aggregation
Membrane proteins have hydrophobic TM helices → aggregate when extracted from membrane
Disordered regions in membrane proteins → especially problematic
Understanding this triangle explains why your purification failed—and how to fix it.
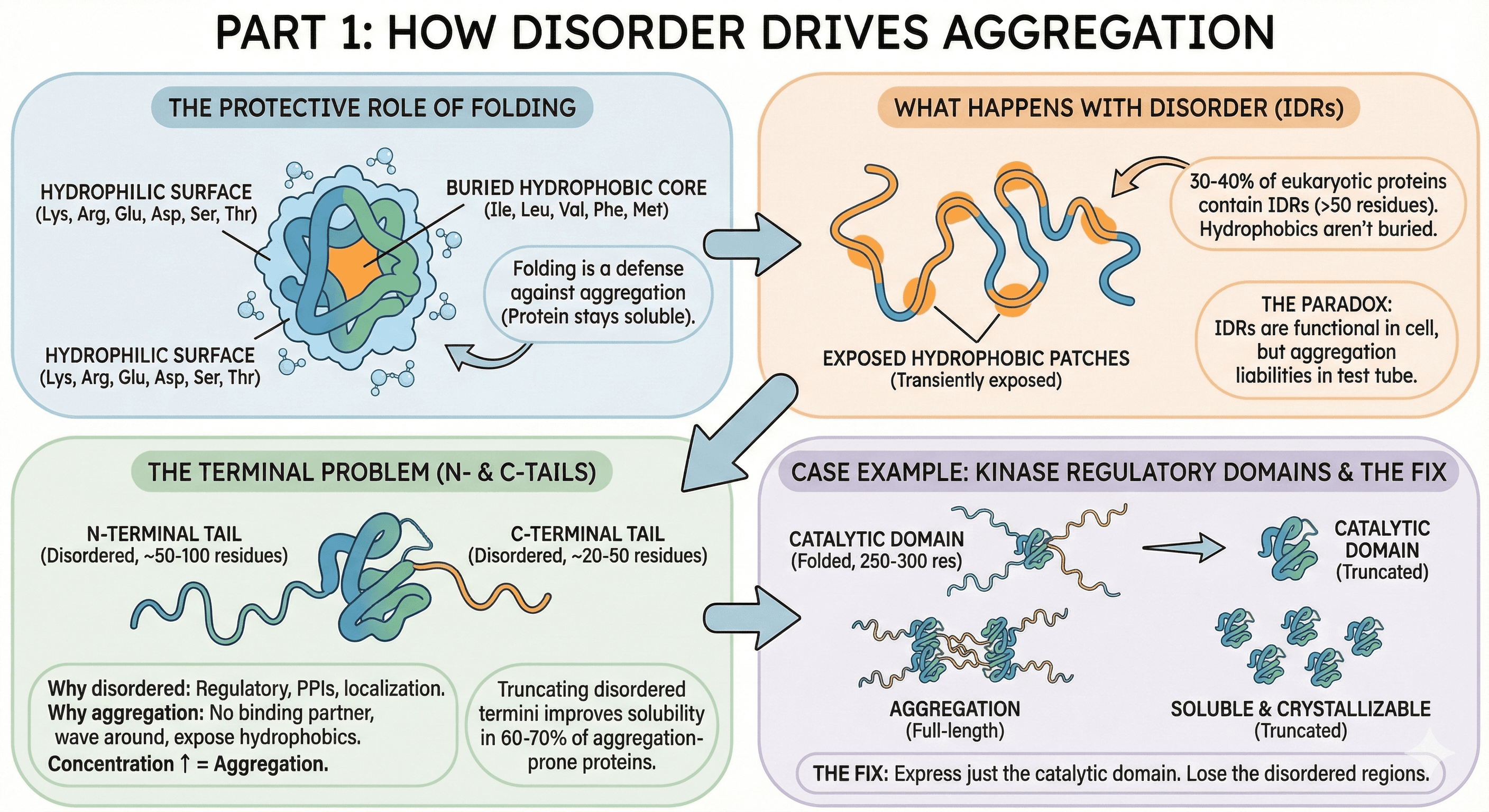
Part 1: How Disorder Drives Aggregation
The Protective Role of Folding
In a well-folded protein:
Hydrophobic residues (Ile, Leu, Val, Phe, Met) are buried in the core
The surface is mostly hydrophilic (Lys, Arg, Glu, Asp, Ser, Thr)
Water interacts with the hydrophilic surface
Protein stays soluble
Folding is a defense mechanism against aggregation.
What Happens When Regions Are Disordered
About 30-40% of eukaryotic proteins contain significant disordered regions (>50 residues), and disorder is present in some form in ~70% of all proteins (Dunker et al., 2008). These intrinsically disordered regions (IDRs) don't have stable tertiary structure, which means:
Hydrophobic residues aren't stably buried
They transiently expose to solvent
Exposed hydrophobic patches stick to each other
Aggregation nucleates
The paradox: IDRs are often functional in the cell (protein-protein interactions, signaling, localization). But in a test tube, without their binding partners, they become aggregation liabilities. IDPs are notably overrepresented in protein aggregates associated with neurodegenerative diseases (Vendruscolo, 2022).
The Terminal Problem
The most common disorder-driven aggregation involves N- and C-terminal tails:
Why termini are often disordered:
Termini are frequently regulatory regions
They mediate protein-protein interactions
They contain localization signals
They're evolutionarily flexible
Why this causes aggregation:
No binding partner in your purification buffer
Disordered termini wave around, exposing hydrophobic patches
Concentration increases → more collisions → aggregation
The data:
30-40% of eukaryotic proteins have disordered termini (>20 residues)
Truncating disordered termini improves solubility in 60-70% of aggregation-prone proteins
This is the single most effective intervention for post-purification aggregation
Case Example: Kinase Regulatory Domains
Many kinases have:
Disordered N-terminal region (50-100 residues)
Well-folded catalytic domain (250-300 residues)
Disordered C-terminal tail (20-50 residues)
In the cell:
N-terminus binds regulatory partners
C-terminus contains phosphorylation sites for regulation
Both are functional
In your purification:
N-terminus has no partners → aggregates
C-terminus has no phosphatases → aggregates
Catalytic domain is dragged along
The fix:
Express just the catalytic domain
Lose the disordered regions
Gain a soluble, crystallizable protein
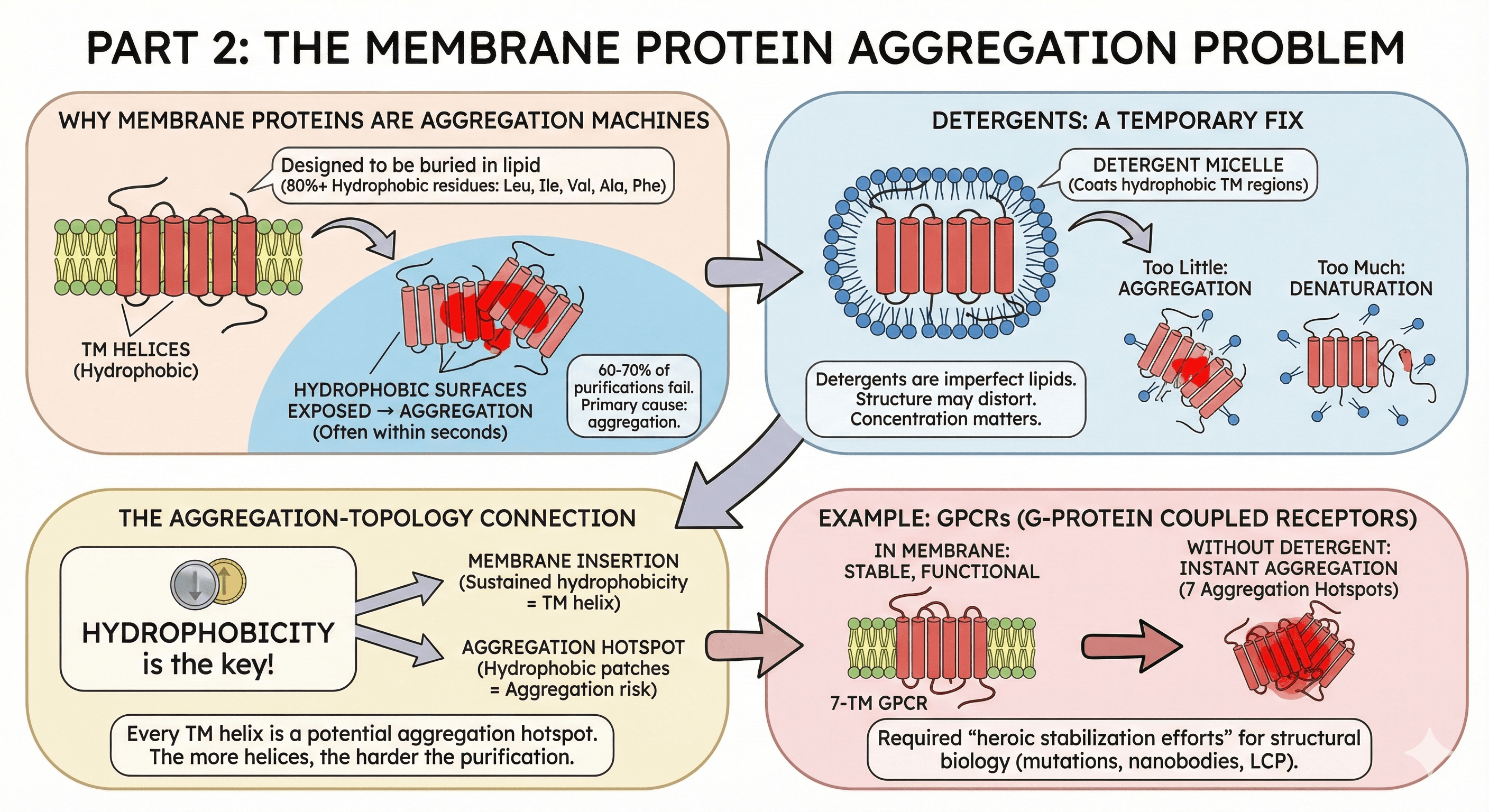
Part 2: The Membrane Protein Aggregation Problem
Why Membrane Proteins Are Aggregation Machines
Transmembrane (TM) helices have a specific job: span the hydrophobic core of the lipid bilayer. To do this, they're composed almost entirely of hydrophobic residues.
A typical TM helix:
20-25 residues long
80%+ hydrophobic (Leu, Ile, Val, Ala, Phe)
Designed to be buried in lipid
What happens during purification:
You lyse cells → membrane is disrupted
TM helices are exposed to aqueous buffer
Hydrophobic surfaces find each other
Aggregation—often within seconds
The numbers:
60-70% of membrane protein purifications fail
Primary cause: aggregation during detergent extraction
GPCRs, ion channels, transporters are all affected
Detergents: A Temporary Fix
Detergents solubilize membrane proteins by:
Coating the hydrophobic TM regions
Creating a micelle that mimics the membrane
Keeping the protein in solution
But detergents are imperfect:
They're not lipids—structure may distort
They don't stabilize the same way membranes do
Concentration matters: too little → aggregation; too much → denaturation
Many proteins are unstable even in optimal detergent
The Aggregation-Topology Connection
Here's where it gets interesting: the same residues that make a helix "transmembrane" make it "aggregation-prone."
Prediction tools use hydrophobicity to identify:
TM helices (sustained hydrophobicity = membrane span)
Aggregation hotspots (hydrophobic patches = aggregation risk)
They're detecting the same underlying property from different angles.
This means:
Every TM helix is a potential aggregation hotspot
Membrane proteins have multiple aggregation hotspots (one per TM helix)
The more TM helices, the harder the purification
Example: GPCRs
G-protein coupled receptors have 7 TM helices = 7 aggregation hotspots:
In membrane: stable, functional receptor
In detergent: marginally stable, activity decays over hours
Without detergent: instant aggregation
This is why GPCR structural biology required heroic stabilization efforts (thermostabilizing mutations, nanobody stabilization, lipidic cubic phase crystallization).
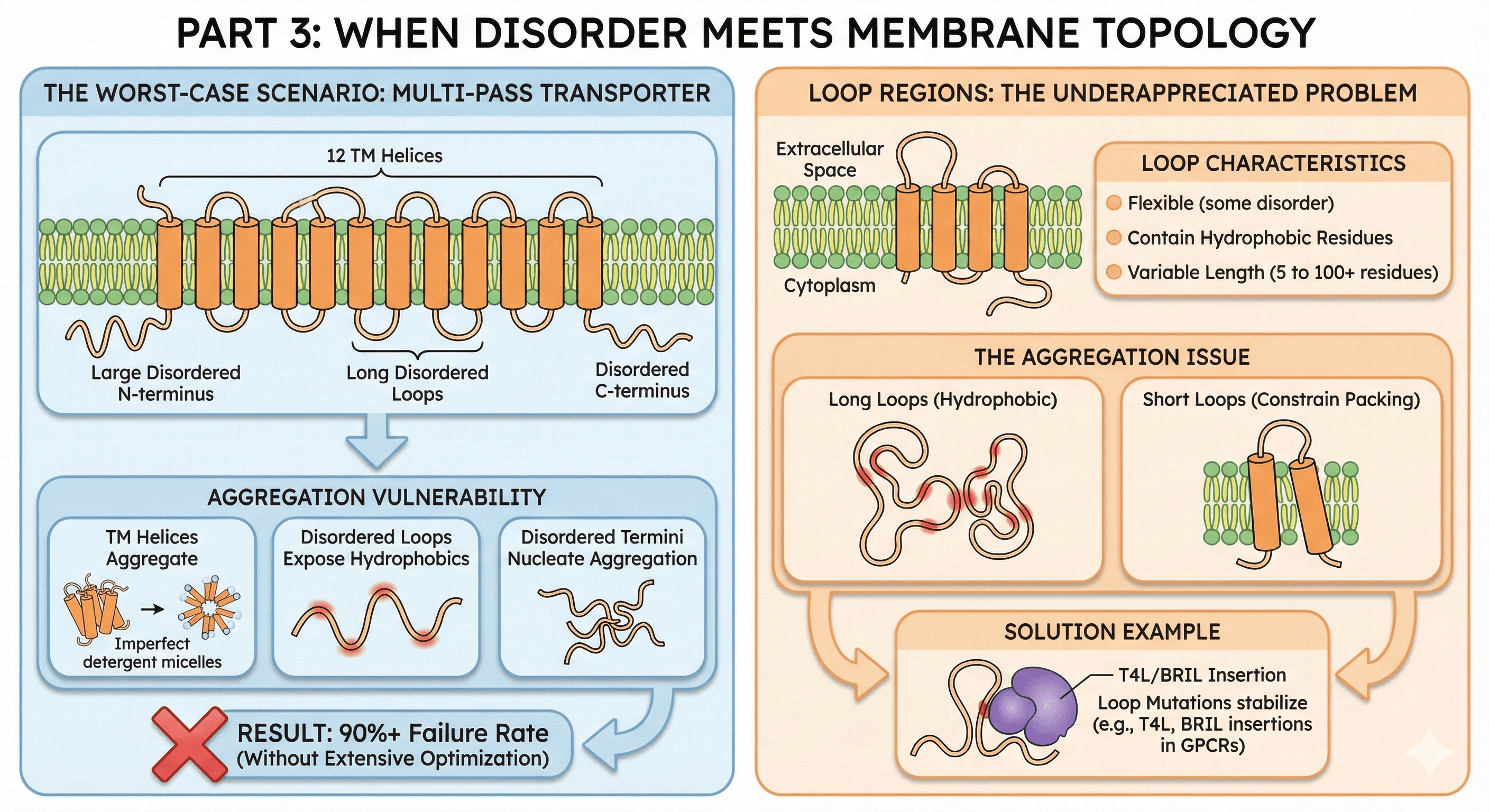
Part 3: When Disorder Meets Membrane Topology
The Worst-Case Scenario
Some proteins have both:
Transmembrane topology
Disordered cytoplasmic loops or termini
Example: Multi-pass transporters
A typical transporter might have:
12 TM helices
Large disordered N-terminus (regulatory)
Long disordered loops between TM helices
Disordered C-terminus
The aggregation vulnerability:
TM helices aggregate if detergent coverage is imperfect
Disordered loops don't fold → expose hydrophobic residues
Disordered termini flail around → nucleate aggregation
Multiple failure modes compound
Result: 90%+ failure rate without extensive optimization
Loop Regions: The Underappreciated Problem
Between TM helices, membrane proteins have loops that face either:
Extracellular space
Cytoplasm
Loop characteristics:
Often flexible (some disorder)
May contain hydrophobic residues (for partner binding or membrane association)
Variable length (from 5 to 100+ residues)
The aggregation issue:
Long loops with hydrophobic character aggregate
Short loops constrain TM helix packing → destabilize the fold
Loop mutations often designed to stabilize (T4L, BRIL insertions in GPCRs)
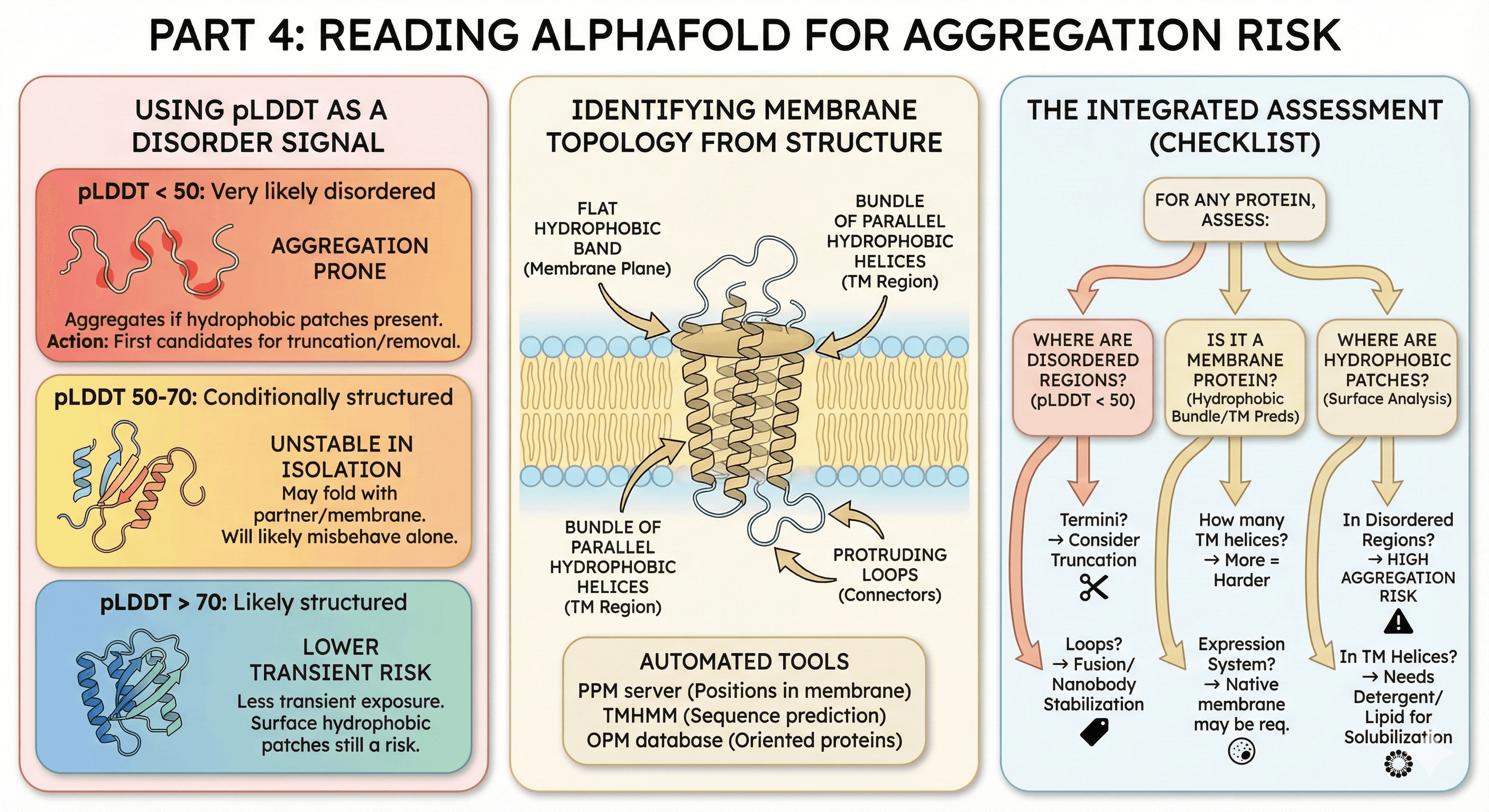
Part 4: Reading AlphaFold for Aggregation Risk
AlphaFold doesn't predict aggregation directly, but it tells you a lot about disorder and topology—and now you know these connect to aggregation.
Using pLDDT as a Disorder Signal
pLDDT < 50: Very likely disordered
These regions will aggregate in vitro if they contain hydrophobic patches
First candidates for truncation or removal
pLDDT 50-70: Conditionally structured
May fold with binding partner
May be stable in membrane context
Will likely misbehave in isolation
pLDDT > 70: Likely structured
Not necessarily aggregation-proof (surface hydrophobic patches still matter)
But less likely to expose hydrophobic residues transiently
Identifying Membrane Topology from Structure
AlphaFold predicts 3D structure, but you need to infer:
Which helices are transmembrane
Where the membrane boundaries are
Which loops are intra- vs extracellular
Visual clues:
Bundle of parallel hydrophobic helices = TM region
Flat "band" of hydrophobic residues = membrane plane
Loops protruding from bundle = connecting loops
Automated tools:
PPM server (positions protein in membrane)
TMHMM on sequence (predicts TM helices)
OPM database (oriented proteins in membrane)
The Integrated Assessment
For any protein, assess:
Where are the disordered regions? (pLDDT < 50)
Termini? → Consider truncation
Loops? → May need fusion proteins or nanobody stabilization
Is it a membrane protein? (Hydrophobic helix bundle, TM predictions)
How many TM helices? → More = harder
What expression system? → Native membrane may be required
Where are the hydrophobic patches? (Surface analysis)
In disordered regions? → High aggregation risk
In TM helices? → Need detergent/lipid for solubilization
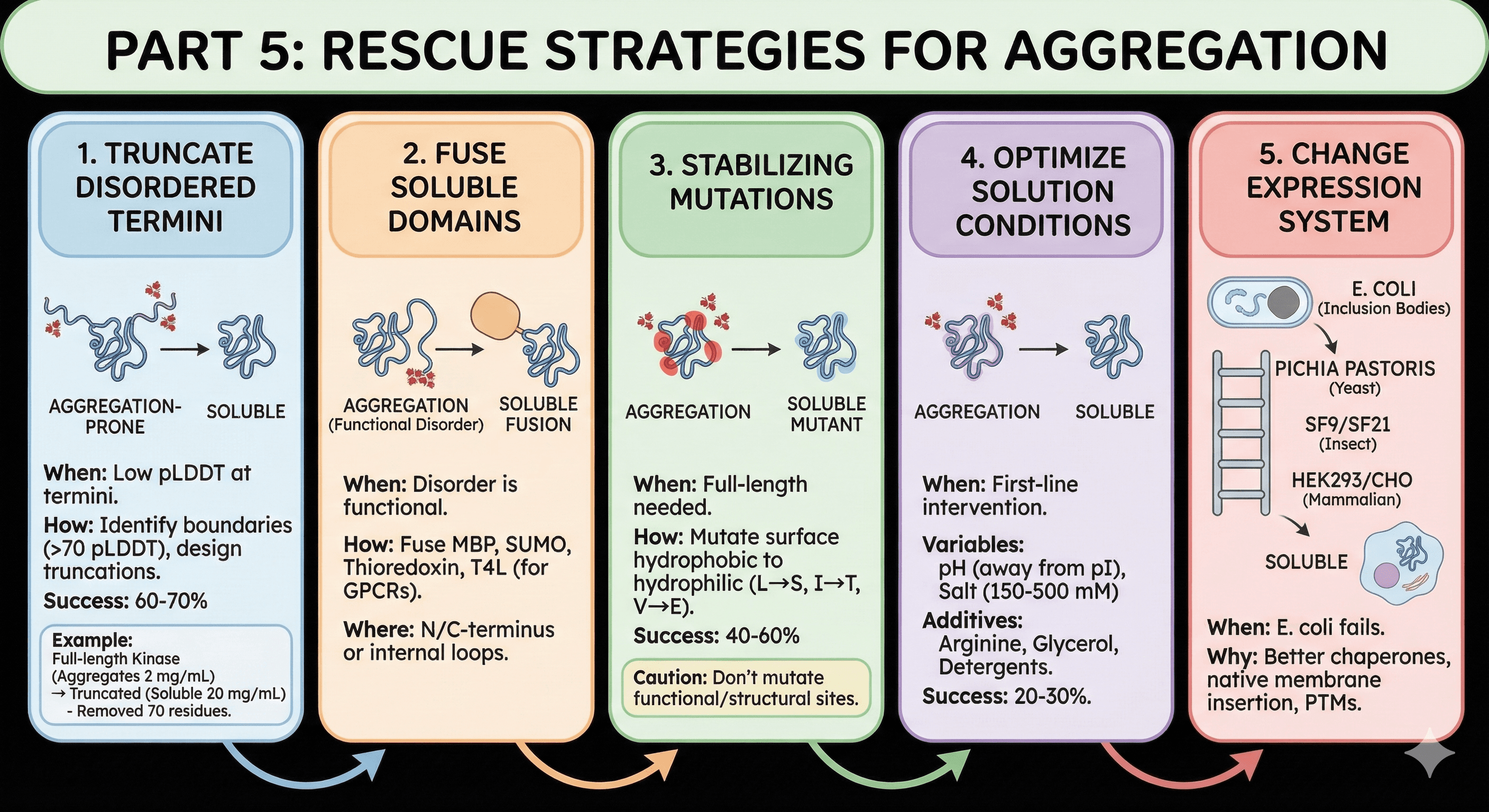
Part 5: Rescue Strategies
Strategy 1: Truncate Disordered Termini
When to use: pLDDT shows low-confidence termini
How to do it:
Identify disorder boundaries (where pLDDT rises above 70)
Design constructs that start/end at structured regions
Make 2-3 truncations at different boundaries
Test expression and solubility
Success rate: 60-70% of aggregation-prone proteins become soluble with terminal truncation
Example:
Full-length kinase (1-450): Aggregates at 2 mg/mL
Truncated kinase (51-420): Soluble at 20 mg/mL
Truncated 70 disordered residues, removed 2 aggregation hotspots
Strategy 2: Fuse Soluble Domains to Mask Aggregation
When to use: Disordered regions are functionally required
Common fusions:
MBP (maltose binding protein): Highly soluble, masks hydrophobic patches
SUMO: Improves folding, clean cleavage
Thioredoxin: Good for small proteins with aggregation issues
T4 Lysozyme (for GPCRs): Stabilizes TM helices by replacing flexible loops
Where to fuse:
N-terminus: Standard, usually least disruptive
C-terminus: If N-terminus is functionally important
Internal (loop replacement): For membrane proteins, replaces disordered loops
Strategy 3: Stabilizing Mutations
When to use: You need the full-length protein but it aggregates
Target selection:
Identify surface hydrophobic residues
Prioritize those in or near disordered regions
Mutate to hydrophilic residues (L→S, I→T, V→E)
Caution:
Don't mutate functional residues
Don't destabilize the fold (check ΔΔG)
Don't remove essential PTM sites
Success rate: 40-60% improvement with 2-3 mutations
Strategy 4: Optimize Solution Conditions
When to use: First-line intervention while planning construct redesign
Variables to screen:
pH: 0.5-1 unit away from pI (reduces aggregation)
Salt: 150-500 mM (increases ionic strength, reduces non-specific interactions)
Additives:
Arginine (100-500 mM): Disrupts hydrophobic interactions
Glycerol (10-20%): Stabilizes native state
Detergents (0.01-0.1%): For membrane proteins
Success rate: 20-30% of aggregating proteins can be rescued with buffer optimization alone
Strategy 5: Change Expression System
When to use: E. coli produces inclusion bodies despite optimization
The expression ladder:
E. coli: If this fails...
Pichia pastoris (yeast): Slower folding, eukaryotic chaperones
Sf9/Sf21 (insect): Better membrane protein handling
HEK293/CHO (mammalian): Native-like folding environment
Why switching helps:
Eukaryotic cells have better chaperone machinery
Membrane protein insertion is native
PTMs that aid folding are present
Case Study: Rescuing a Triple-Threat Protein
The Target
Human transporter protein:
12 TM helices
80-residue disordered N-terminus
Long disordered loop between TM6 and TM7
Critical for drug transport—needed for pharmacology studies
Initial Attempt
Expression: Sf9 insect cells Purification: DDM extraction, His-tag purification Result: Protein extracted, but aggregated within 4 hours at 4°C
Diagnosis
Analysis revealed:
Disorder: N-terminus (1-80) and loop (residues 320-380) show pLDDT < 40
Aggregation hotspots: Both disordered regions contain hydrophobic patches
TM instability: 12 TM helices require constant detergent coverage
The triple threat: Disorder + membrane topology + aggregation propensity
The Solution
Step 1: Truncate N-terminus
Removed residues 1-75
Kept 5 residues before first TM helix
Step 2: Replace disordered loop
Inserted T4 lysozyme (T4L) between TM6 and TM7
Rigid insertion stabilizes TM packing
Standard GPCR engineering strategy applied to transporter
Step 3: Optimize detergent
DDM → LMNG (longer chain, better coverage)
Added CHS (cholesterol hemisuccinate) for membrane stability
Step 4: Add nanobody
Raised nanobody against stable conformation
Nanobody binding further stabilizes the fold
The Result
Before: 4-hour stability, aggregates at 2 mg/mL
After: 48-hour stability, soluble at 5 mg/mL
Cryo-EM structure: Solved at 3.2 Å resolution
Key insight: All three problems (disorder, topology, aggregation) were addressed together. Fixing only one wouldn't have worked.
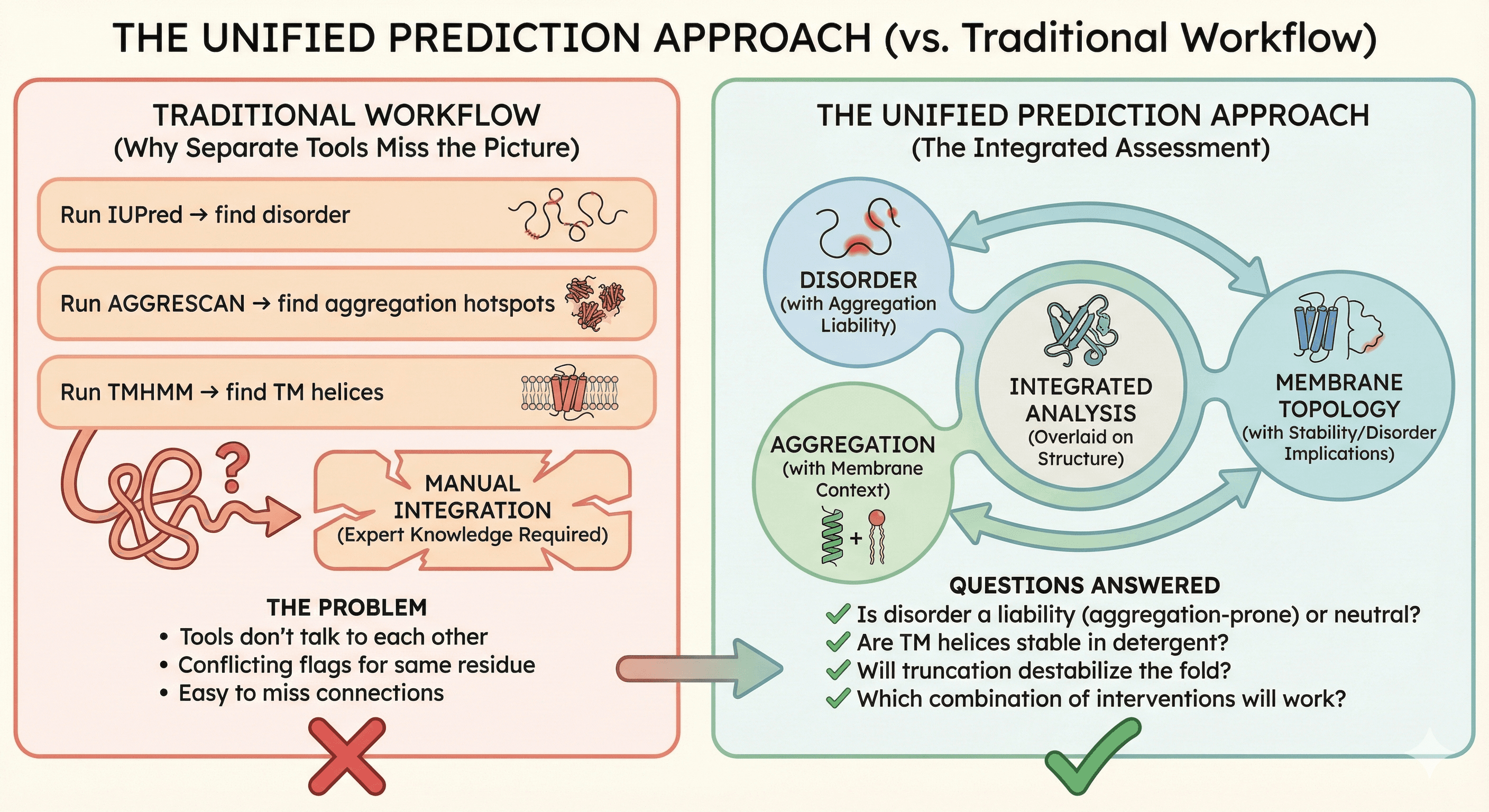
The Unified Prediction Approach
Why Separate Tools Miss the Picture
Traditional workflow:
Run IUPred → find disorder
Run AGGRESCAN → find aggregation hotspots
Run TMHMM → find TM helices
Try to integrate manually
The problem:
Tools don't talk to each other
Same residue might be flagged differently by each tool
Integration requires expert knowledge
Easy to miss connections
The Integrated Assessment
What you actually need:
Disorder prediction that considers aggregation consequences
Aggregation prediction that accounts for membrane context
Membrane topology with disorder and stability implications
All three overlaid on the structure
Questions an integrated analysis answers:
Is this disordered region a liability (aggregation-prone) or neutral (hydrophilic)?
Are these TM helices stable in detergent, or do they need specific lipids?
Will truncating this terminus destabilize the fold?
Which combination of interventions will work?
Practical Decision Tree
The Bottom Line
Disorder, aggregation, and membrane topology aren't three separate problems. They're manifestations of the same underlying biophysics: hydrophobic residues and their exposure to water.
Property | What It Means | Consequence |
|---|---|---|
Disorder | No stable structure | Hydrophobic residues transiently exposed |
Aggregation propensity | Hydrophobic surface patches | Proteins stick to each other |
Membrane topology | Hydrophobic TM helices | Aggregate outside membrane |
The unified principle: Anything that exposes hydrophobic residues to water is an aggregation risk.
Understanding this connection transforms troubleshooting:
Not "my protein is disordered" but "my protein exposes aggregation hotspots"
Not "my protein needs detergent" but "my TM helices aggregate without membrane mimetics"
Not "three separate problems" but "one problem with three manifestations"
Unified Analysis for Aggregation Rescue
For researchers dealing with aggregation-prone proteins, platforms like Orbion provide integrated analysis of disorder, aggregation propensity, and membrane topology from a single prediction. This unified view reveals:
Which disordered regions are actually aggregation liabilities
How membrane topology contributes to purification difficulty
Where to truncate, mutate, or fuse to rescue your protein
What expression system and purification conditions to prioritize
The goal is to see the connections between these properties before you start purifying—so you can design constructs that avoid the aggregation trap entirely, rather than troubleshooting it for months afterward.
References
Dunker AK, et al. (2008). Function and structure of inherently disordered proteins. Current Opinion in Structural Biology, 18(6):756-764. PMC2443096
Vendruscolo M, et al. (2022). Intrinsically disordered proteins identified in the aggregate proteome serve as biomarkers of neurodegeneration. Frontiers in Neuroscience, 15:780567. PMC8748380
Uversky VN. (2013). Intrinsic disorder-based protein interactions and their modulators. Current Pharmaceutical Design, 19(23):4191-4213. Link
Chiti F & Dobson CM. (2017). Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annual Review of Biochemistry, 86:27-68. Link
Krogh A, et al. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of Molecular Biology, 305(3):567-580. Link
Fernandez-Escamilla AM, et al. (2004). Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nature Biotechnology, 22(10):1302-1306. Link

