Blog
Why Stabilizing Mutations Sometimes Make Everything Worse
Jan 28, 2026
You ran the stability prediction software. It suggested L134I would increase thermal stability by 3°C. You made the mutation, expressed the protein, measured the Tm, and celebrated: 47°C to 50°C, just as predicted. Then you ran the activity assay. The enzyme was dead.
Welcome to the stability-function tradeoff—the reason that "stabilizing mutations" can destroy your protein.
Key Takeaways
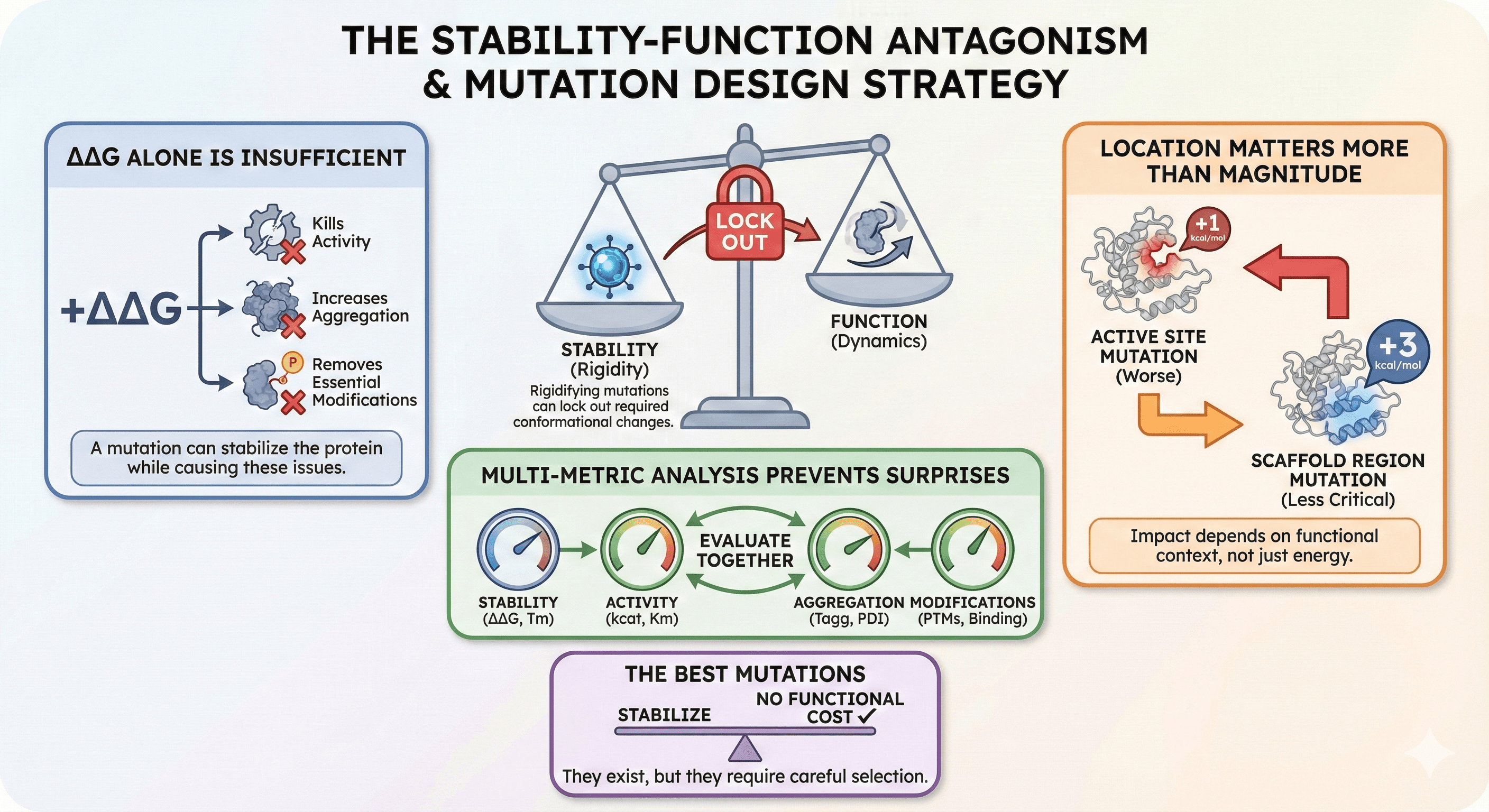
Stability and function are often antagonistic: Mutations that rigidify the fold can lock out required conformational changes
ΔΔG alone is insufficient: A mutation can stabilize the protein while killing activity, increasing aggregation, or removing essential modifications
Location matters more than magnitude: A +1 kcal/mol mutation in the active site is worse than a +3 kcal/mol mutation in a scaffold region
Multi-metric analysis prevents surprises: Evaluate stability, activity, aggregation, and modifications together
The best mutations stabilize without functional cost: They exist, but they require careful selection
The Stability-Function Paradox
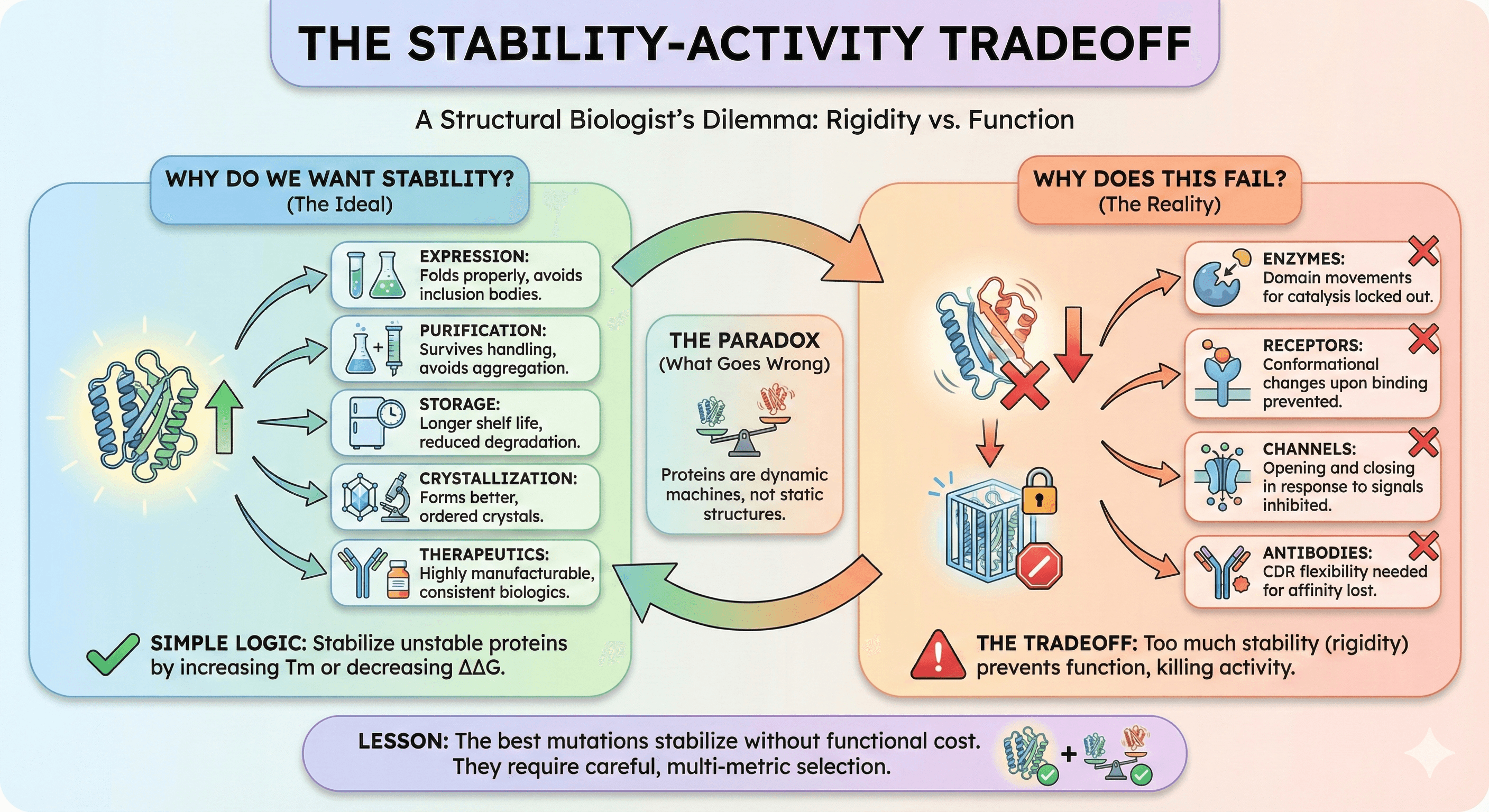
Why Do We Want Stability?
Stable proteins are easier to work with:
Expression: Stable proteins fold properly, avoiding inclusion bodies
Purification: Stable proteins survive handling without aggregating
Storage: Stable proteins have longer shelf life
Crystallization: Stable proteins form better crystals
Therapeutics: Stable biologics are more manufacturable
The logic seems simple: If the protein is unstable, stabilize it. Find mutations that increase Tm or decrease ΔΔG, make the changes, problem solved.
Why Does This Fail?
Proteins aren't static structures. They're dynamic machines that function through motion (Henzler-Wildman & Kern, 2007):
Enzymes require domain movements for catalysis
Receptors undergo conformational changes upon ligand binding
Channels open and close in response to signals
Antibodies need CDR flexibility for affinity maturation
Stabilizing mutations reduce dynamics. That's the whole point—you're making the protein more rigid. But if the protein needs flexibility to work, rigidifying it kills function. This stability-activity tradeoff is well-documented across enzyme families (Tokuriki et al., 2008).
The paradox: Stability is good, but too much stability prevents function.
The Five Ways Stabilizing Mutations Go Wrong
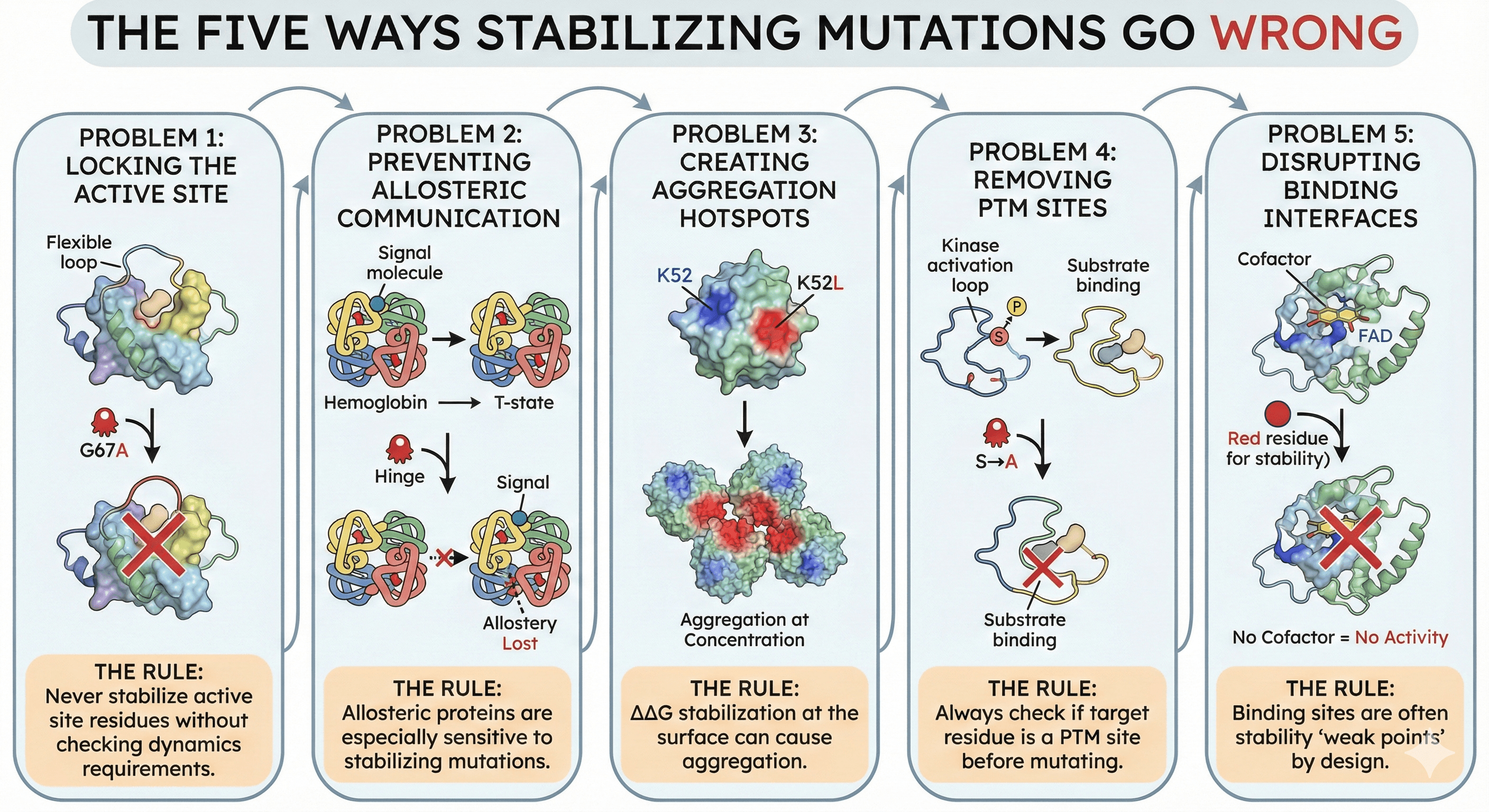
Problem 1: Locking the Active Site
The scenario:
Enzyme has flexible active site loop that opens to accept substrate
Mutation rigidifies the loop (increases Tm)
Loop can no longer open
Substrate can't enter
Activity: zero
Example: Lysozyme
Wild-type lysozyme:
Active site has mobile loop (residues 65-75)
Loop opens to accommodate polysaccharide substrate
Closes for catalysis
Stabilizing mutation (hypothetical: G67A):
Removes glycine flexibility
Loop can't open
Substrate excluded
Tm +2°C, activity -90%
The rule: Never stabilize active site residues without checking dynamics requirements.
Problem 2: Preventing Allosteric Communication
The scenario:
Protein has allosteric regulation
Binding at one site affects conformation at another
Stabilizing mutation breaks the conformational link
Allostery lost
Example: Hemoglobin
Hemoglobin's cooperative oxygen binding depends on:
Conformational change at one subunit
Transmitted to other subunits
T-state to R-state transition
A mutation that over-stabilizes T-state:
Blocks transition to R-state
Destroys cooperativity
Now binds oxygen poorly
The rule: Allosteric proteins are especially sensitive to stabilizing mutations.
Problem 3: Creating Aggregation Hotspots
The scenario:
Mutation adds hydrophobicity for stability (e.g., K→L)
The new hydrophobic residue is surface-exposed
Creates an aggregation-prone patch
Protein is more stable but aggregates at concentration
The perverse outcome: Tm increases, but practical solubility decreases.
Example: Antibody engineering
Original residue: K52 (charged, soluble) Stabilizing mutation: K52L (hydrophobic core, ΔΔG = -0.8 kcal/mol)
Result:
Individual Fab is more stable
But surface hydrophobic patch promotes aggregation
At therapeutic concentrations (>100 mg/mL): precipitates
The rule: ΔΔG stabilization at the surface can cause aggregation.
Problem 4: Removing PTM Sites
The scenario:
Residue is a PTM site (phosphorylation, glycosylation)
The modification is essential for function or localization
Mutation for stability removes the modifiable residue
Protein loses its regulatory mechanism
Example: Kinase activation loop
Many kinases have activation loop phosphorylation:
Unphosphorylated: Low activity (autoinhibited)
Phosphorylated: High activity (active)
Stabilizing mutation that removes phosphorylation site:
Can lock kinase in either state
If locked inactive: Dead enzyme
If locked active: Potential oncogene
Real case: Mutations at kinase activation loops are frequently oncogenic. Some "stabilizing" mutations cause cancer.
The rule: Always check if target residue is a PTM site before mutating.
Problem 5: Disrupting Binding Interfaces
The scenario:
Protein interacts with partners (substrates, cofactors, other proteins)
Interface residues are sometimes suboptimal for stability
Mutating them for stability disrupts binding
Protein is more stable but can't do its job
Example: Enzyme-cofactor interaction
Many enzymes bind cofactors (NAD, FAD, heme, metal ions):
Cofactor binding site has specific geometry
Residues evolved for binding, not for protein stability
These residues might look "improvable" to stability predictors
Mutation at cofactor site:
May stabilize the apo protein
But disrupts cofactor binding
No cofactor = no activity
The rule: Binding sites are often stability "weak points" by design.
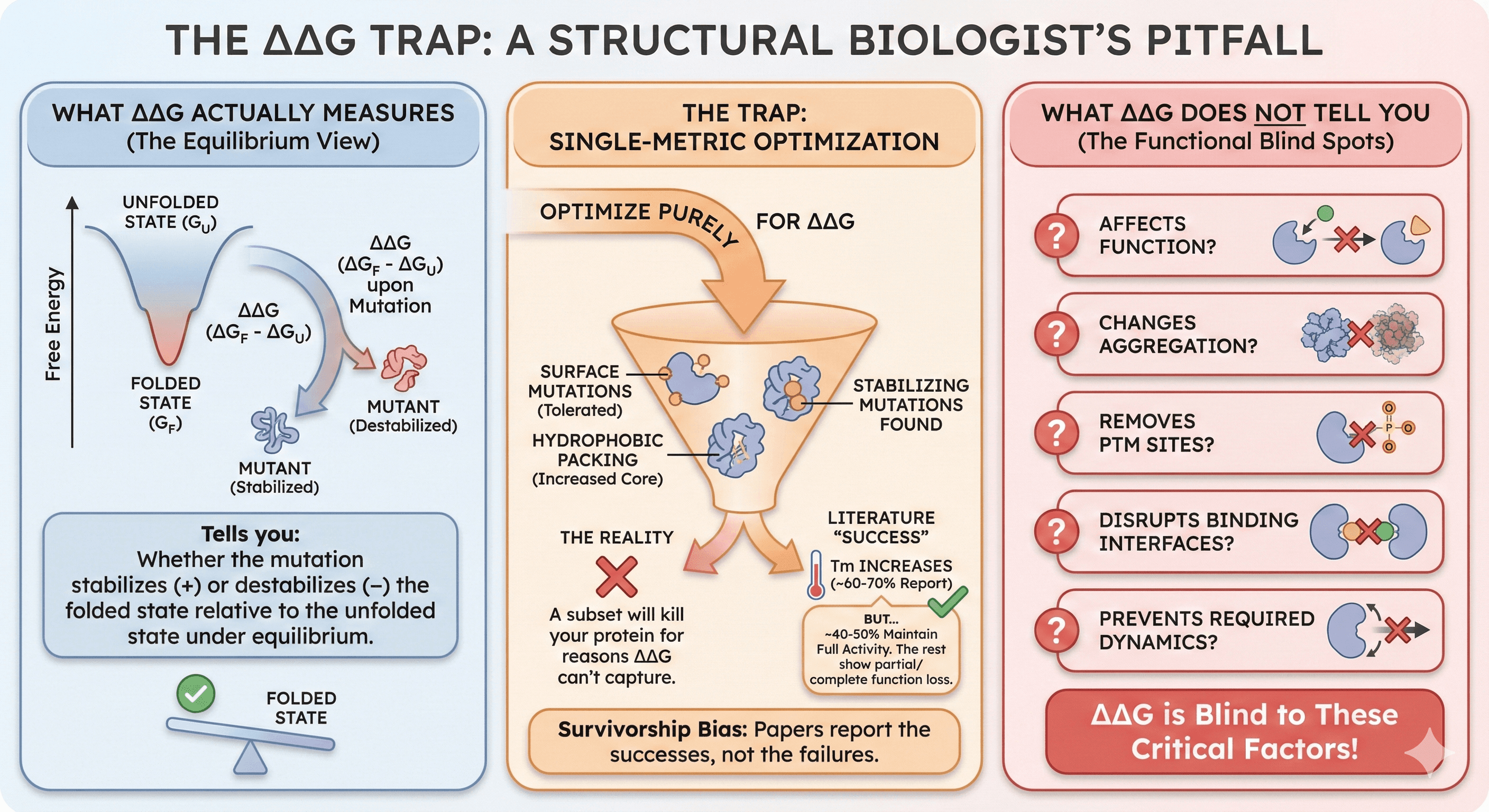
The ΔΔG Trap
What ΔΔG Actually Measures
ΔΔG (change in Gibbs free energy upon mutation) tells you:
Whether the mutation stabilizes (+) or destabilizes (-) the folded state
Relative to the unfolded state
Under equilibrium conditions
What ΔΔG does NOT tell you:
Whether the mutation affects function
Whether it changes aggregation propensity
Whether it removes PTM sites
Whether it disrupts binding interfaces
Whether it prevents required dynamics
Why Single-Metric Optimization Fails
If you optimize purely for ΔΔG:
You'll find stabilizing mutations
Many will be on the protein surface (where mutations are most tolerated)
Some will add hydrophobicity (increasing core packing)
A subset will kill your protein for reasons ΔΔG can't capture
The numbers:
Literature reports ~60-70% success rate for ΔΔG-predicted stabilizing mutations
"Success" means Tm increases
But only ~40-50% maintain full activity
The rest show partial or complete function loss
Survivorship bias: Papers report the mutations that worked. The failures don't get published.
Multi-Metric Mutation Analysis
The Correct Approach
Evaluate each candidate mutation across multiple dimensions:
Metric | What It Tells You | Red Flag |
|---|---|---|
ΔΔG | Fold stability | Destabilizing (< -1 kcal/mol) |
ΔTm | Thermal stability | Significant decrease |
Δ Activity | Function | Any decrease |
Δ Aggregation | Solubility | Increase |
Δ Binding | Partners/cofactors | Disruption |
PTM site | Modifications | Removal |
Δ Disorder | Local flexibility | Wrong direction |
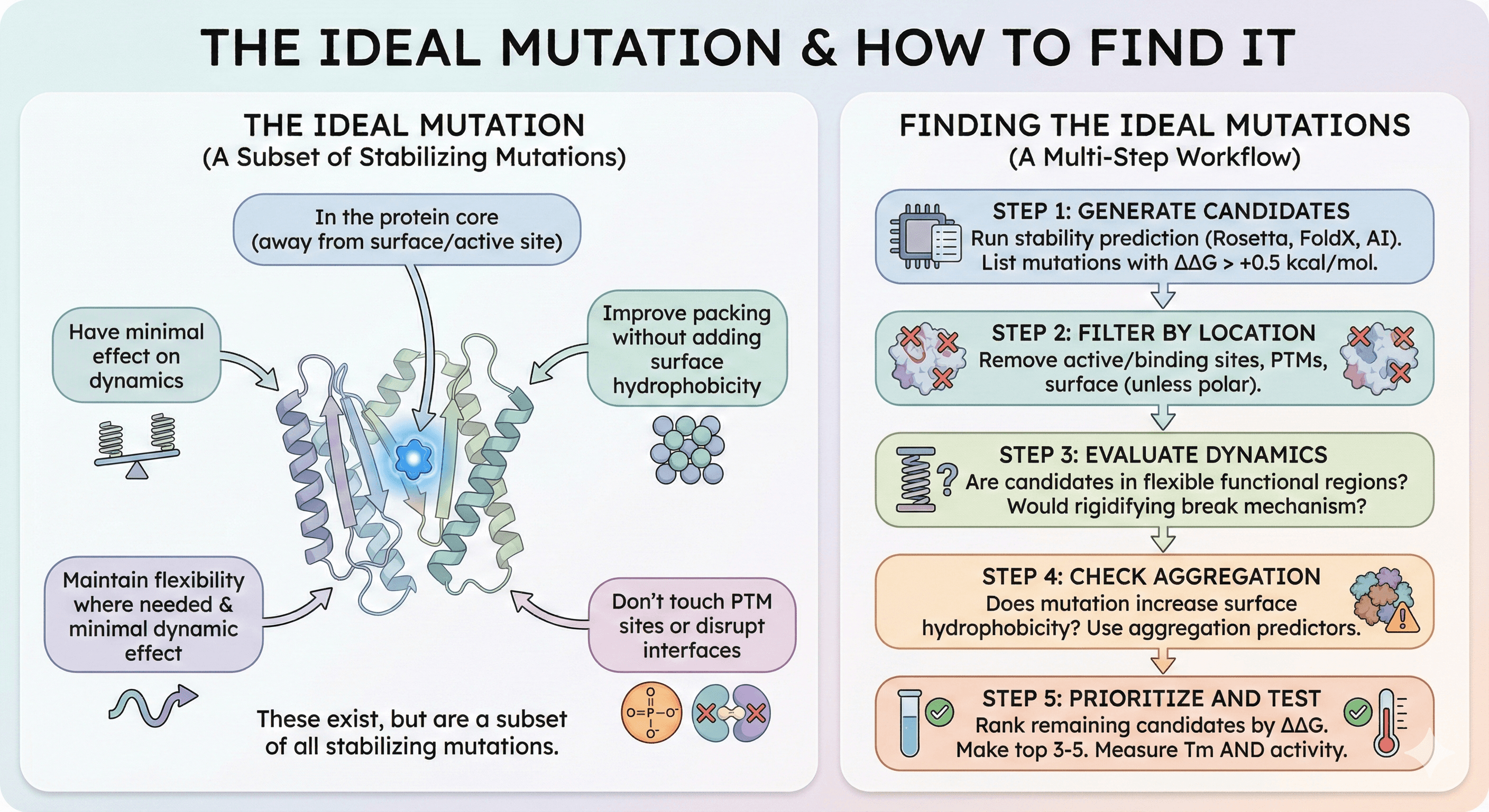
The Ideal Mutation
The best stabilizing mutations are:
In the protein core (away from surface and active site)
Improve packing without adding surface hydrophobicity
Maintain or increase local flexibility where needed
Don't touch PTM sites
Don't disrupt interfaces
Have minimal effect on dynamics
These exist, but they're a subset of all stabilizing mutations.
Finding the Ideal Mutations
Step 1: Generate candidates
Run stability prediction (Rosetta, FoldX, or AI tools)
List all mutations with ΔΔG > +0.5 kcal/mol
Step 2: Filter by location
Remove active site residues
Remove known binding site residues
Remove PTM sites
Remove surface residues (unless switching to charged/polar)
Step 3: Evaluate dynamics
Are any candidates in flexible regions required for function?
Would rigidifying this region break the mechanism?
Step 4: Check aggregation
Does the mutation increase surface hydrophobicity?
Use aggregation predictors on mutant sequence
Step 5: Prioritize and test
Rank remaining candidates by ΔΔG
Make top 3-5 mutations
Measure Tm AND activity
Case Studies in Mutation Tradeoffs
Case 1: The Dead Enzyme
Target: Metabolic enzyme for biochemical assay Problem: Enzyme loses activity within 24 hours at 4°C Goal: Stabilize for longer shelf life
Approach 1 (failed):
Ran Rosetta ΔΔG prediction
Top mutation: F67W (core packing improvement)
Made mutation, measured Tm: +4°C
Measured activity: 5% of wild-type
F67 is adjacent to active site; larger Trp blocks substrate entry
Approach 2 (successful):
Filtered Rosetta results to exclude active site region
Top mutation outside active site: V198I (distant from catalytic center)
Made mutation, measured Tm: +2°C
Measured activity: 100% of wild-type
Shelf life: Extended from 24 hours to 1 week
Lesson: Exclude the functional machinery from stability optimization.
Case 2: The Aggregating Antibody
Target: Therapeutic antibody for chronic disease Problem: Aggregates above 50 mg/mL (need 150 mg/mL for subcutaneous) Goal: Stabilize to prevent aggregation
Approach 1 (failed):
Identified flexible CDR loop with low Tm
Rigidified loop with V31I (ΔΔG = +1.1 kcal/mol)
Tm increased from 62°C to 66°C
Aggregation unchanged
Binding affinity dropped 10-fold
Approach 2 (successful):
Analyzed aggregation propensity, not just stability
Identified surface hydrophobic patch (I53, F55)
Mutated to polar: I53S, F55T (ΔΔG = +0.3 kcal/mol only)
Tm slightly increased (63°C)
Aggregation threshold: 50 mg/mL → 180 mg/mL
Binding affinity: maintained
Lesson: Aggregation and stability are different problems requiring different solutions.
Case 3: The Unregulated Kinase
Target: Kinase for drug discovery (need stable protein for crystallography) Problem: Kinase is unstable, Tm = 42°C, can't crystallize Goal: Increase Tm for structural studies
Approach 1 (dangerous):
Predicted stabilizing mutation T201V (in activation loop)
T201 is a phosphorylation site
Mutation locks kinase in partially active state
Tm increases to 52°C (stable!)
But: This is now a different conformational state
Crystal structure would not represent physiological state
Approach 2 (correct):
Identified stabilizing mutations outside regulatory regions
Mutations in core helices: L87I, A145V
Tm increased to 50°C
Activation loop still functional
Phosphorylation still possible
Crystal structure represents regulatable kinase
Lesson: Don't confuse "stable" with "correct state."
The Right Way to Stabilize
Step-by-Step Workflow
What to Optimize (And What Not To)
Good targets for stabilization:
Core hydrophobic residues (packing improvements)
Buried polar residues (hydrogen bond networks)
Surface residues (changing to charged/polar)
Loop regions (if not functionally required)
Bad targets for stabilization:
Active site (catalysis)
Binding site (substrates, cofactors, partners)
PTM sites (regulation)
Flexible regions required for mechanism
Interface residues (if function requires binding)
The Economics of Getting It Wrong
Time and Money
If you pick the wrong mutation:
Express mutant protein: 1-2 weeks
Purify and characterize: 1 week
Discover function is compromised: Day 1 of assays
Redesign and try again: Add 3-4 weeks
If you pick the right mutation first:
Same expression and purification: 2-3 weeks
Function confirmed: Day 1 of assays
Move forward with stable, functional protein
Difference: 3-4 weeks and the demoralizing experience of solving the wrong problem.
The Therapeutic Context
For biologics development:
A destabilizing mutation discovered in clinical trials = program termination
An aggregation-prone variant = formulation failure = manufacturing rebuild
Loss of PTM site = altered pharmacokinetics = potentially dangerous
The cost of wrong mutations in pharma: Millions of dollars and years of delay.
Tools for Multi-Metric Analysis
What's Available
Traditional stability predictors (ΔΔG only):
Rosetta: Gold standard, but complex
FoldX: Faster, widely used
MAESTRO: Quick empirical predictions
Limitations: Only predict fold stability, not function or other properties.
Aggregation predictors:
AGGRESCAN: Sequence-based hotspot detection
CamSol: Solubility prediction
AGGRESCAN3D: Structure-based
Limitations: Don't connect to stability or function.
What's missing: Integrated platforms that evaluate all dimensions simultaneously.
The Ideal Analysis
For each candidate mutation, you want to see:
ΔΔG: Is this actually stabilizing?
Δ Disorder: Am I rigidifying a region that needs flexibility?
Δ Aggregation: Am I creating a surface hydrophobic patch?
PTM impact: Am I removing a modification site?
Binding site check: Am I in or near a functional interface?
pLDDT change: Does the local structure prediction change?
The result: A mutation score that captures the full picture, not just one dimension.
The Bottom Line
Stability optimization isn't about maximizing ΔΔG. It's about finding the subset of stabilizing mutations that don't break anything else.
Metric Alone | Risk | Better Approach |
|---|---|---|
ΔΔG only | Functional loss | ΔΔG + location filter |
Tm only | Aggregation | Tm + aggregation |
Surface hydrophobicity | Aggregation | Balance with polar |
Any single metric | Missing the full picture | Multi-metric |
The paradox resolved: Stability and function can coexist—but only if you select mutations that are stabilizing in regions that don't matter for function.
The difference between a successful stabilization project and a failed one is usually one thing: checking more than just ΔΔG.
Comprehensive Variant Analysis
For researchers engineering protein stability, platforms like Orbion provide multi-dimensional variant analysis that goes beyond single-metric predictions:
Thermodynamic metrics: ΔTm, ΔΔG predictions
Structural/biophysical impacts: Changes in disorder, aggregation propensity, local confidence
Functional preservation: Binding site analysis, PTM site detection
Residue-level comparison: See exactly what changes at each position
The goal is to find stabilizing mutations that preserve function—not to blindly maximize stability and hope nothing breaks.
References
Henzler-Wildman K & Kern D. (2007). Dynamic personalities of proteins. Nature, 450:964-972. Link
Tokuriki N, et al. (2008). How protein stability and new functions trade off. PLoS Computational Biology, 4(2):e1000002. PMC2904692
Schymkowitz J, et al. (2005). The FoldX web server: an online force field. Nucleic Acids Research, 33(Web Server issue):W382-W388. Link
Kellogg EH, et al. (2011). Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins, 79(3):830-838. Link
Bloom JD, et al. (2006). Protein stability promotes evolvability. PNAS, 103(15):5869-5874. Link
Wang X, et al. (2018). Antibody structure, instability, and formulation. Journal of Pharmaceutical Sciences, 96(1):1-26. Link
Ó Conchúir S, et al. (2015). A web resource for standardized benchmark datasets, metrics, and Rosetta protocols for macromolecular modeling and design. PLoS One, 10(9):e0130433. Link