Blog
How to Choose Expression Systems for Your Protein's PTM Requirements
Dec 2, 2025
The fundamental question in protein engineering: Which expression system should you use? The answer isn't about convenience—it's about post-translational modifications (PTMs).
Post-translational modifications determine whether your protein will work. They're not decorative—they're functional. PTMs affect folding, stability, localization, and activity. Get them wrong, and your beautifully designed construct becomes an expensive pile of misfolded aggregates.
Key Takeaways
Start simple: Use E. coli unless you have a specific PTM requirement (glycosylation, complex disulfides)
Match biology to biology: Don't use mammalian cells for proteins that don't need eukaryotic PTMs
Know your protein first: Experimental PTM data (UniProt) has gaps—use AI prediction for complete coverage
The cost ladder: E. coli (1×) → Yeast (3-5×) → Insect (10-20×) → Mammalian (20-50×)
Critical decision point: Does your protein require glycosylation for folding? If yes, skip E. coli
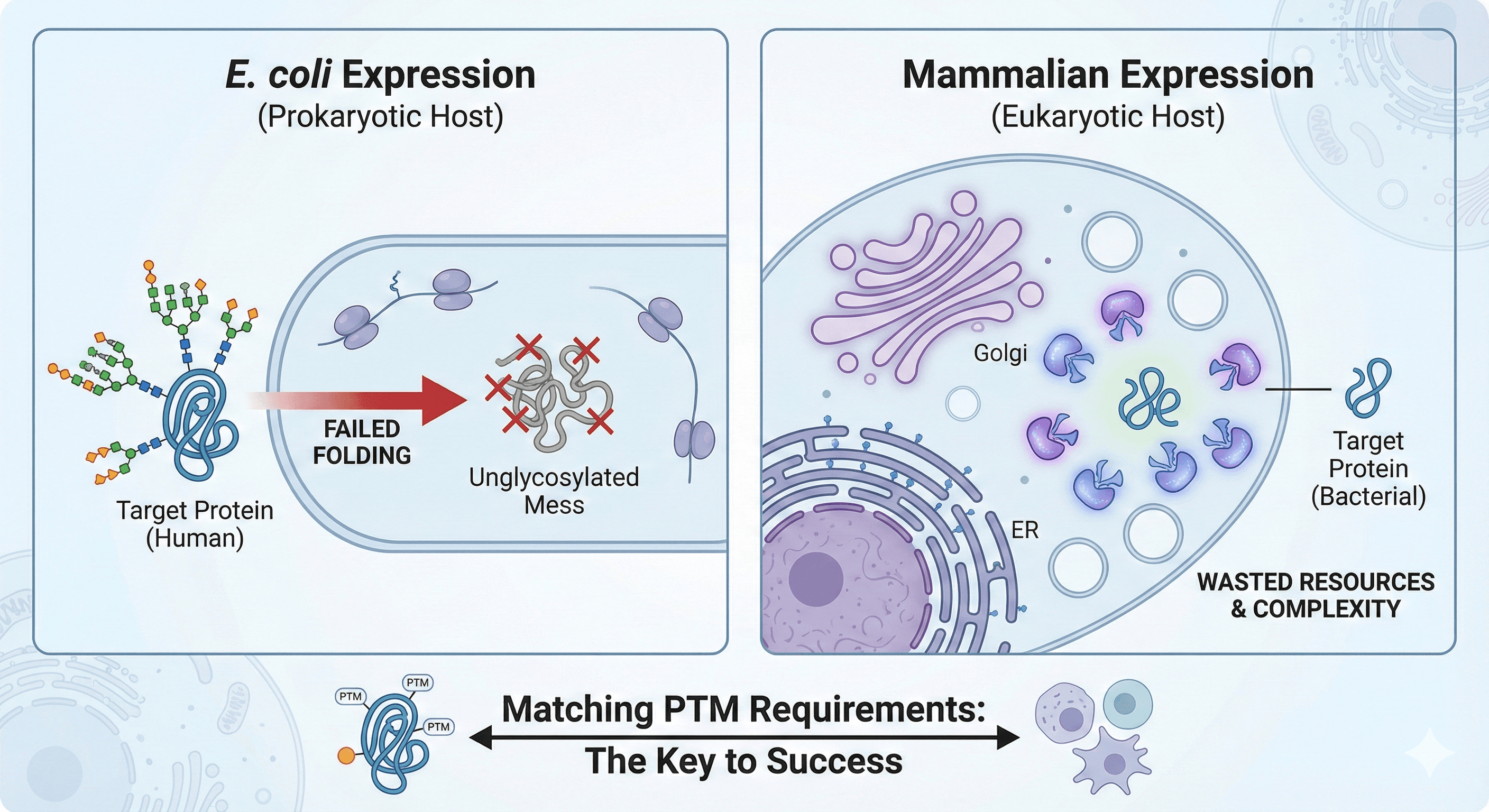
But here's the problem: not all expression systems can produce all PTMs. Express a heavily glycosylated human protein in E. coli, and you'll get an unglycosylated mess that likely won't fold. Express a simple bacterial protein in mammalian cells, and you're wasting time and money on unnecessary complexity.
Choosing the right expression system means matching your protein's PTM requirements to what the host cell can actually deliver. Let me walk you through how to make that decision.
Quick Comparison: Expression Systems at a Glance
System | Timeline | Cost | Yield | Best For | Deal Breaker |
|---|---|---|---|---|---|
E. coli | 1-2 days | $ | 1-10 g/L | Cytoplasmic proteins, no glycosylation | Needs N-glycosylation |
Yeast | 3-5 days | $$$ | 0.1-1 g/L | Secreted proteins, disulfides | Needs human glycans |
Insect | 5-7 days | $$$$ | 1-10 mg/L | Membrane proteins, GPCRs | Needs full sialylation |
Mammalian | 10-14 days | $$$$$ | 0.05-5 g/L | Therapeutics, human PTMs | High cost/low throughput |
The Expression System Ladder
Think of expression systems as a ladder. You want to start at the bottom (fastest, cheapest) and only climb higher when you need to.
E. coli: Fast and Cheap, But Limited
Timeline: 24-48 hours from transformation to protein
Cost: $$ (baseline)
Yield: 1-10 g/L for well-expressing proteins
Best for:
Cytoplasmic proteins without disulfide bonds
Proteins that don't require eukaryotic PTMs
High-throughput screening (need to test 50+ variants quickly)
Budget-constrained academic projects
Enzymes, structural domains, fluorescent proteins
What E. coli can do:
Phosphorylation (limited - mostly His/Asp on two-component systems; minimal Ser/Thr/Tyr)
Methylation (PrmA, PrmB, PrmC methyltransferases for ribosomal proteins)
Acetylation (both enzymatic via Pat/AcP and non-enzymatic)
Some lipidation (N-terminal myristoylation possible with NMT expression; no eukaryotic palmitoylation)
What E. coli cannot do:
N-glycosylation (no machinery)
O-glycosylation (no machinery)
Complex disulfide bonds (cytoplasm is reducing environment)
Tyrosine sulfation
Most eukaryotic PTMs
Common failures:
Membrane proteins (aggregate without proper folding)
Antibodies (require disulfide bonds, need periplasmic expression or refolding)
Secreted human proteins (usually need glycosylation to fold)
When it works: You're making interleukins, simple enzymes, fluorescent proteins, structural domains without complex PTMs. E. coli is your workhorse. Fast, cheap, scalable.
When it fails: You need glycosylation, you're working with a GPCR, or you're making a therapeutic antibody. Don't waste weeks trying to make E. coli work for a protein it fundamentally can't handle.

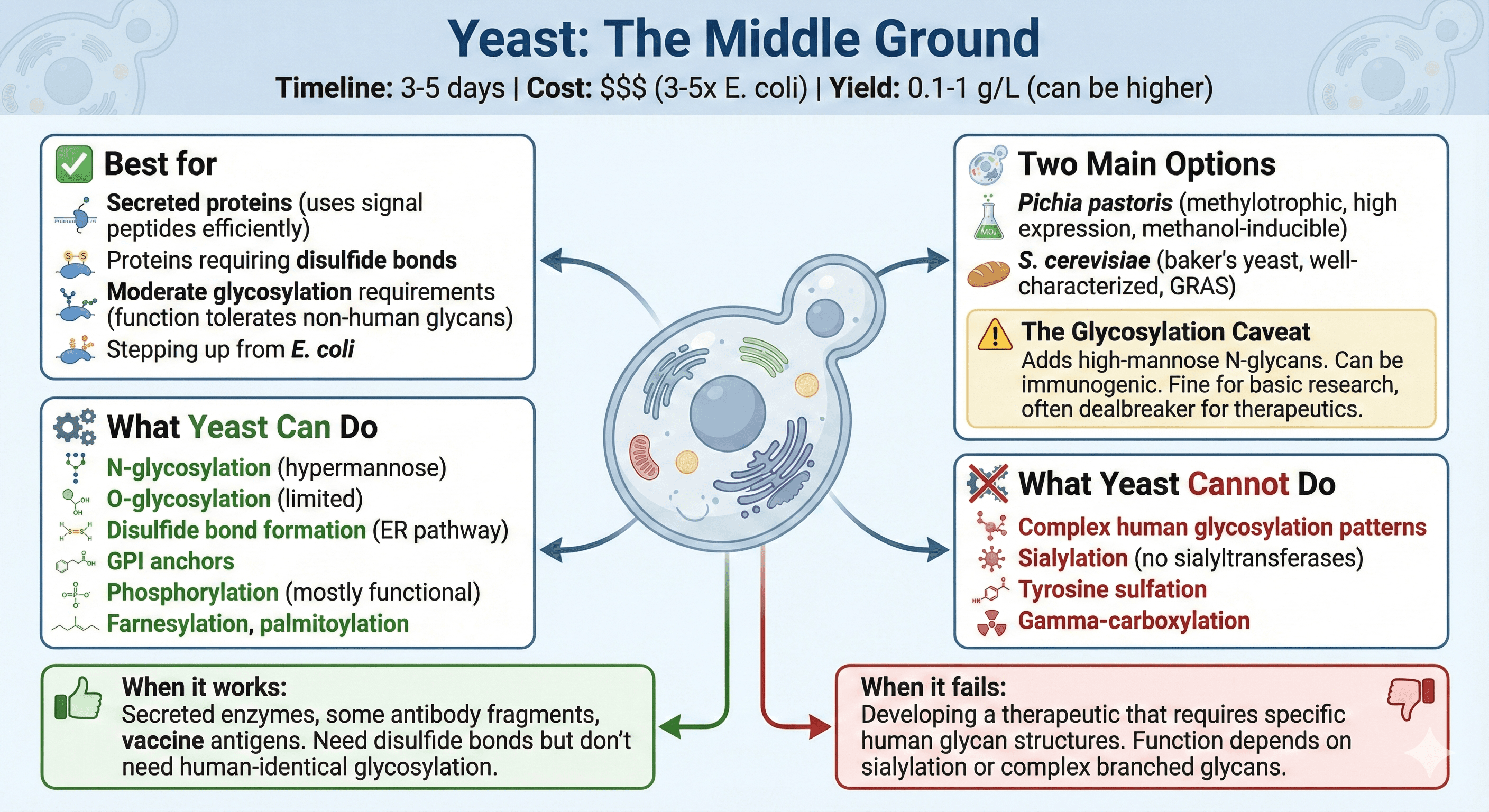
Yeast: The Middle Ground
Timeline: 3-5 days from transformation to protein
Cost: $$$ (3-5× E. coli)
Yield: 0.1-1 g/L (can reach higher with optimization)
Two main options:
Pichia pastoris (methylotrophic yeast, high expression, methanol-inducible)
S. cerevisiae (baker's yeast, well-characterized, GRAS status for therapeutics)
Best for:
Secreted proteins (uses signal peptides efficiently)
Proteins requiring disulfide bonds
Moderate glycosylation requirements (function tolerates non-human glycans)
Stepping up from E. coli without going full mammalian
What yeast can do:
N-glycosylation (but hypermannose, not human-like)
O-glycosylation (limited)
Disulfide bond formation (ER pathway)
GPI anchors
Phosphorylation (mostly functional)
Farnesylation, palmitoylation
What yeast cannot do:
Complex human glycosylation patterns
Sialylation (no sialyltransferases)
Tyrosine sulfation
Gamma-carboxylation
The glycosylation caveat: Yeast glycosylates, but differently. It adds high-mannose N-glycans that can be immunogenic for therapeutics. For basic research, this often doesn't matter. For drug development, it's a dealbreaker.
When it works: You're expressing secreted enzymes, some antibody fragments, vaccine antigens. You need disulfide bonds but don't need human-identical glycosylation.
When it fails: You're developing a therapeutic that requires specific human glycan structures. Your protein's function depends on sialylation or complex branched glycans.
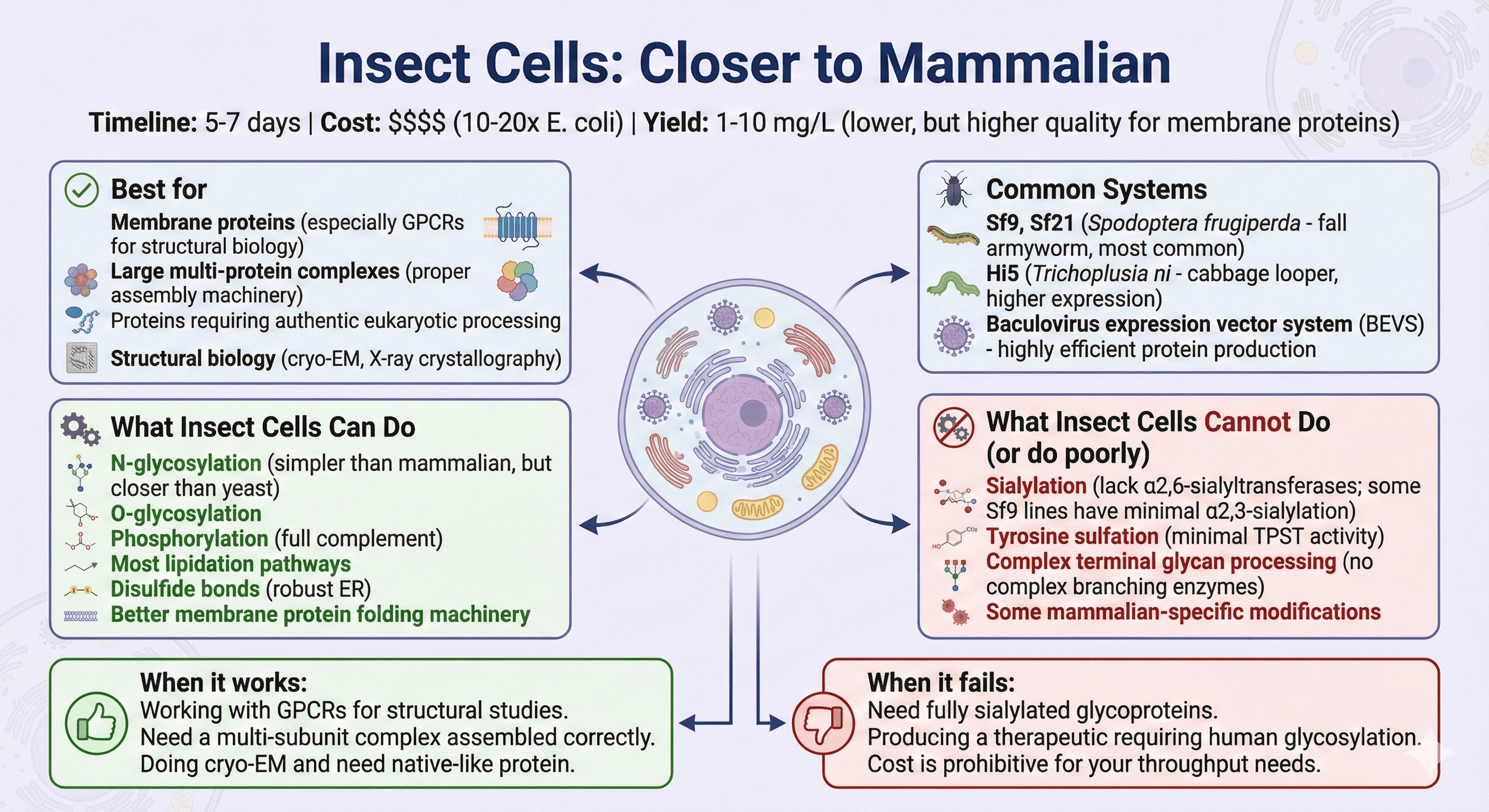
Insect Cells: Closer to Mammalian
Timeline: 5-7 days from transfection to protein
Cost: $$$$ (10-20× E. coli)
Yield: 1-10 mg/L (lower than E. coli, but higher quality for membrane proteins)
Common systems:
Sf9, Sf21 (Spodoptera frugiperda - fall armyworm, most common)
Hi5 (Trichoplusia ni - cabbage looper, higher expression)
Baculovirus expression vector system (BEVS) - highly efficient protein production
Best for:
Membrane proteins (especially GPCRs for structural biology)
Large multi-protein complexes (proper assembly machinery)
Proteins requiring authentic eukaryotic processing
Structural biology (cryo-EM, X-ray crystallography)
What insect cells can do:
N-glycosylation (simpler than mammalian, but closer than yeast)
O-glycosylation
Phosphorylation (full complement)
Most lipidation pathways
Disulfide bonds (robust ER)
Better membrane protein folding machinery
What insect cells cannot do (or do poorly):
Sialylation (lack α2,6-sialyltransferases; some Sf9 lines have minimal α2,3-sialylation)
Tyrosine sulfation (minimal TPST activity)
Complex terminal glycan processing (no complex branching enzymes)
Some mammalian-specific modifications
When it works: You're working with GPCRs for structural studies. You need a multi-subunit complex assembled correctly. You're doing cryo-EM and need native-like protein.
When it fails: You need fully sialylated glycoproteins. You're producing a therapeutic requiring human glycosylation. Cost is prohibitive for your throughput needs.
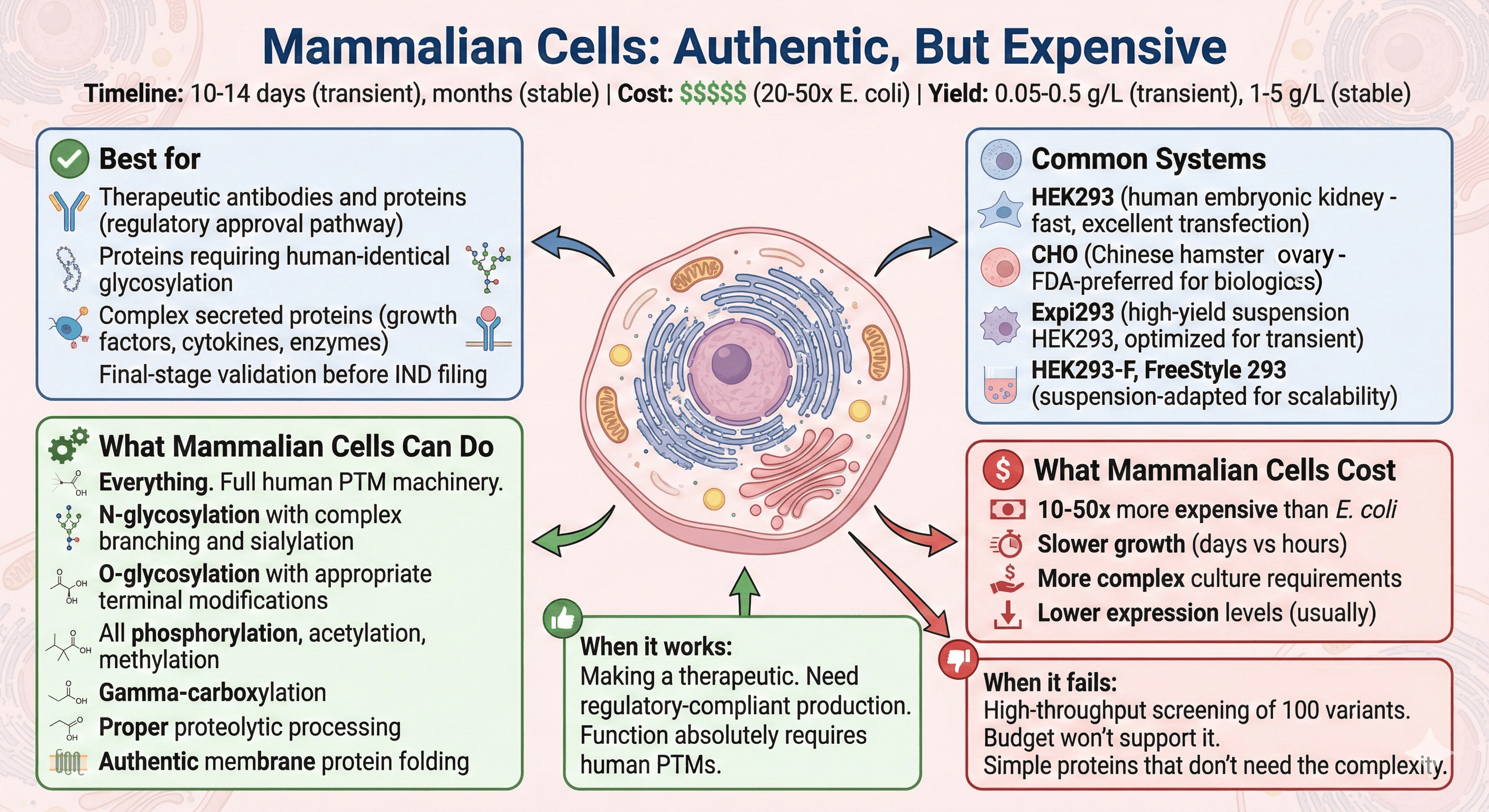
Mammalian Cells: Authentic, But Expensive
Timeline: 10-14 days for transient expression; months for stable cell lines
Cost: $$$$$ (20-50× E. coli)
Yield: 0.05-0.5 g/L transient; 1-5 g/L stable lines (CHO can reach higher)
Common systems:
HEK293 (human embryonic kidney - fast growth, excellent transfection, research gold standard)
CHO (Chinese hamster ovary - FDA-preferred for biologics, ~70% of therapeutic proteins)
Expi293 (high-yield suspension HEK293, optimized for transient expression)
HEK293-F, FreeStyle 293 (suspension-adapted for scalability)
Best for:
Therapeutic antibodies and proteins (regulatory approval pathway)
Proteins requiring human-identical glycosylation
Complex secreted proteins (growth factors, cytokines, enzymes)
Final-stage validation before IND filing
What mammalian cells can do:
Everything. Full human PTM machinery.
N-glycosylation with complex branching and sialylation
O-glycosylation with appropriate terminal modifications
All phosphorylation, acetylation, methylation
Gamma-carboxylation
Proper proteolytic processing
Authentic membrane protein folding
What mammalian cells cost:
10-50x more expensive than E. coli
Slower growth (days vs hours)
More complex culture requirements
Lower expression levels (usually)
When it works: You're making a therapeutic. You need regulatory-compliant production. Function absolutely requires human PTMs.
When it fails: You're doing high-throughput screening of 100 variants. Budget won't support it. Simple proteins that don't need the complexity.
The PTM × Expression System Matrix
Here's the reference table. Bookmark this.
PTM Type | E. coli | Yeast | Insect | Mammalian |
|---|---|---|---|---|
N-glycosylation | ✗ | ✓ (hypermannose) | ✓ (partial) | ✓ (full) |
O-glycosylation | ✗ | △ (limited) | ✓ | ✓ |
Phosphorylation (S/T/Y) | △ (limited) | ✓ | ✓ | ✓ |
Disulfide bonds | △ (periplasm only) | ✓ | ✓ | ✓ |
Acetylation | ✓ | ✓ | ✓ | ✓ |
Methylation | ✓ | ✓ | ✓ | ✓ |
Ubiquitination | ✗ | ✓ | ✓ | ✓ |
Sumoylation | ✗ | ✓ | ✓ | ✓ |
Palmitoylation | ✗ | ✓ | ✓ | ✓ |
Myristoylation | ✗ | ✓ | ✓ | ✓ |
GPI anchor | ✗ | ✓ | ✓ | ✓ |
Sialylation | ✗ | ✗ | △ (weak) | ✓ |
Sulfation | ✗ | ✗ | △ (weak) | ✓ |
Gamma-carboxylation | ✗ | ✗ | ✗ | ✓ |
Legend:
✓ = Robustly supported
△ = Partially supported / limited capacity
✗ = Not supported / minimal activity
The Decision Framework: 3 Steps to Choose Your Expression System
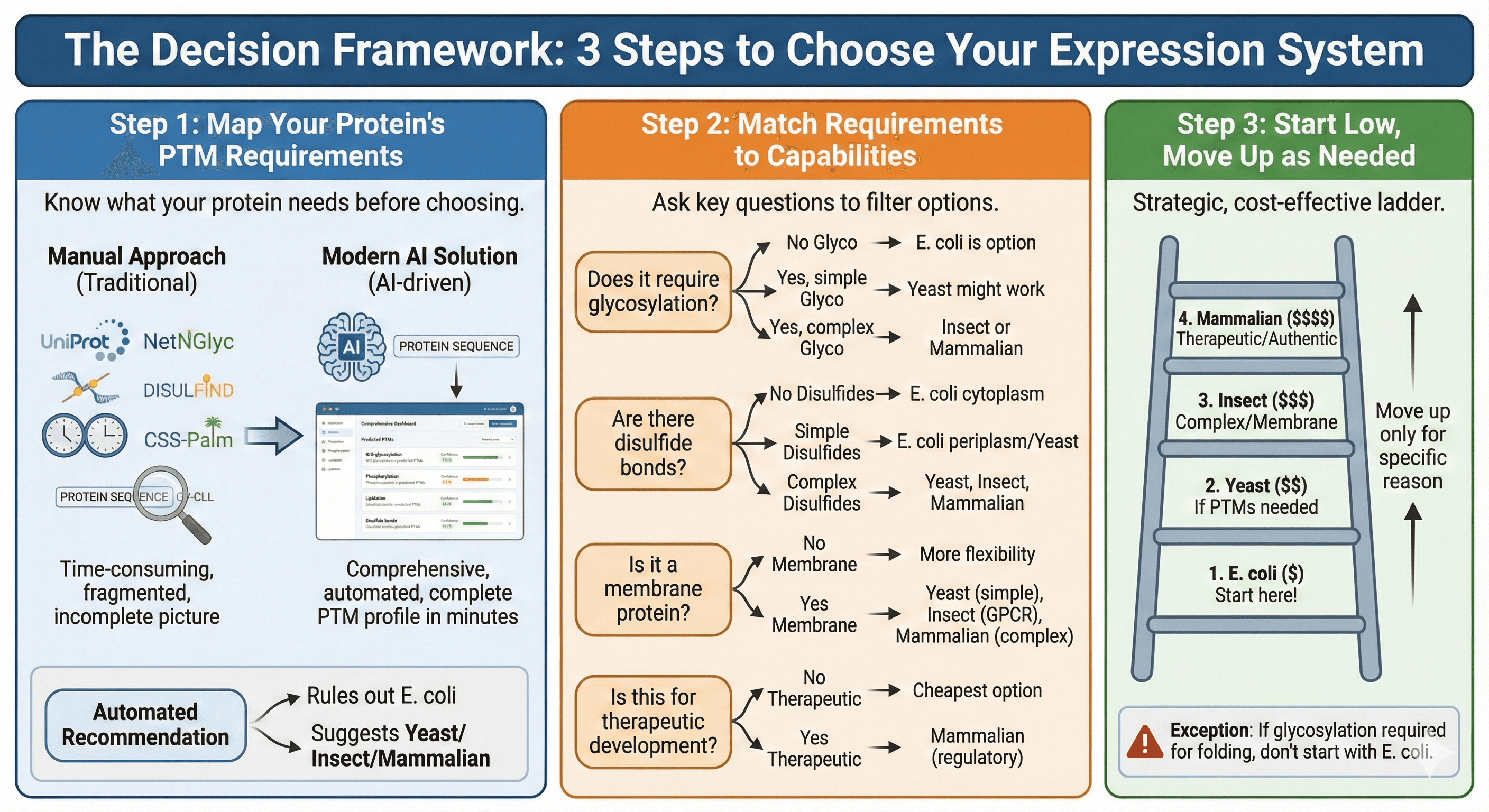
Step 1: Map Your Protein's PTM Requirements
Before you choose a system, know what your protein actually needs.
The Manual Approach (Traditional Workflow)
Sequence analysis:
UniProt annotations - experimentally validated PTMs when available (often incomplete)
NetNGlyc 1.0 - predict N-glycosylation sites (Asn-X-Ser/Thr motifs)
NetOGlyc 4.0 - predict O-glycosylation sites
DISULFIND / DiANNA - predict disulfide bonds from sequence
CSS-Palm 4.0 - predict palmitoylation sites
Structural analysis:
Check PDB for homologous structures showing disulfide bonds
Look for membrane-spanning regions (transmembrane helices = expression challenge)
Identify disordered regions (may need tags for stability)
The problem with this approach:
UniProt has gaps: Most proteins lack comprehensive experimental PTM annotations
Fragmented tools: You need 5+ different prediction servers with different formats
Time-consuming: Checking databases + running predictions takes 1-2 hours per protein
Incomplete picture: Experimental data captures what's been studied, not what exists
The Modern AI Solution
Use machine learning models trained on structural and sequence data to predict the complete PTM landscape, even when experimental data is absent. AI models can infer PTMs from evolutionary patterns, structural context, and sequence motifs—often with higher coverage than databases alone.
How this works in practice (using Orbion's Characterize module as an example):
Input sequence → AI analyzes structural features and evolutionary patterns
Comprehensive PTM prediction:
N- and O-glycosylation sites (beyond simple consensus motifs)
Phosphorylation sites (Ser/Thr/Tyr) with confidence scores
Lipidation sites (palmitoylation, myristoylation, prenylation)
Disulfide bonds from structural context
Membrane topology, signal peptides, domain boundaries
Automated expression system recommendation:
Glycosylation required? → Rules out E. coli
Complex disulfides? → Suggests yeast or higher
Membrane protein? → Recommends insect or mammalian
Therapeutic context? → Prioritizes CHO/HEK293
This gives you a complete, actionable PTM profile in minutes—not the hours required for manual database searches.
Step 2: Match Requirements to Capabilities
Ask these questions:
Does it require glycosylation?
No → E. coli is on the table
Yes, simple → Yeast might work
Yes, complex → Insect or mammalian
Are there disulfide bonds?
No → E. coli cytoplasm is fine
Yes, simple (1-2 bonds) → E. coli periplasm or yeast
Yes, complex → Yeast, insect, or mammalian
Is it a membrane protein?
No → More flexibility
Yes → Yeast (if simple), insect (if GPCR), mammalian (if complex)
Is this for therapeutic development?
No → Use cheapest option that works
Yes → Probably need mammalian (regulatory considerations)
Step 3: Start Low, Move Up as Needed
The default strategy: Start with E. coli. Only move up the ladder when you have a specific reason.
Don't skip steps based on assumptions. Yes, it's a mammalian protein. But maybe it'll express in E. coli anyway. Try before you commit to expensive systems.
Exception: If the protein requires glycosylation for folding (common for secreted proteins), don't waste time on E. coli. Start with yeast or higher.
Common Mistakes
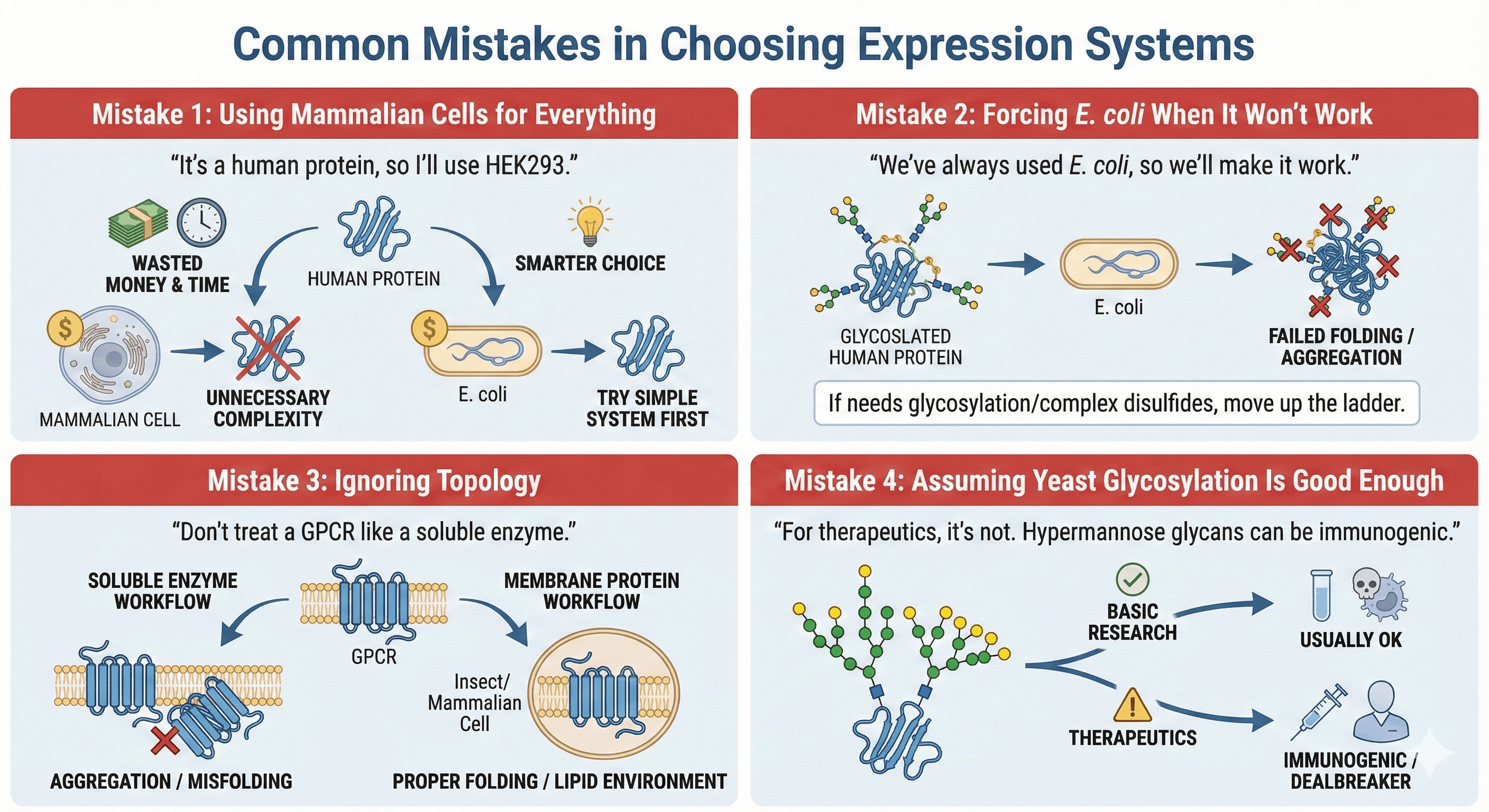
Mistake 1: Using Mammalian Cells for Everything
"It's a human protein, so I'll use HEK293."
This wastes time and money. Many human proteins express perfectly in E. coli. Try the simple system first unless you have a specific PTM requirement.
Mistake 2: Forcing E. coli When It Won't Work
"We've always used E. coli, so we'll make it work."
If your protein needs glycosylation or has complex disulfide bonds, no amount of codon optimization or tag engineering will make E. coli work. Move up the ladder.
Mistake 3: Ignoring Topology
Membrane proteins are special. Transmembrane regions, lipid requirements, proper folding machinery - these matter. Don't treat a GPCR like a soluble enzyme.
Mistake 4: Assuming Yeast Glycosylation Is Good Enough
For basic research, it usually is. For therapeutics, it's not. Hypermannose glycans can be immunogenic. Know the difference.
Real-World Examples
Example 1: Green Fluorescent Protein (GFP)
PTMs needed: None
System choice: E. coli
Why it works: Simple, cytoplasmic, self-folding
Expression level: Very high (grams per liter)
Example 2: β2-Adrenergic Receptor (GPCR)
PTMs needed: Palmitoylation (Cys341), phosphorylation (multiple Ser/Thr in C-terminus), N-glycosylation (Asn6, Asn15)
System choice: Insect cells (Sf9 with baculovirus)
Why E. coli fails: Seven-transmembrane protein, needs ER insertion machinery, lipid environment
Key modification: Often use T4-lysozyme fusion (ICL3 replacement) for crystallization
Expression level: 1-5 mg/L (lower yield, but functional and properly folded)
Example 3: Monoclonal Antibody (IgG)
PTMs needed: 16 disulfide bonds (12 intrachain, 4 interchain), N-glycosylation at Asn297 (Fc region)
System choice: CHO cells (industry standard for >70% of therapeutic mAbs)
Why yeast fails: Glycan structure affects FcγR binding, ADCC, CDC; hypermannose glycans are immunogenic
Critical factor: Glycosylation at Asn297 modulates effector function—must be human-compatible
Expression level: 3-5 g/L in fed-batch CHO; 10+ g/L in optimized perfusion systems
Example 4: Insulin
PTMs needed: Disulfide bonds, proteolytic processing
System choice: E. coli (surprisingly!)
How: Express as inclusion bodies, refold, process in vitro
Why it works: Small, well-characterized refolding protocol, cost-effective at scale
The Bottom Line: Match Biology to Biology
Expression system choice isn't about prestige or default assumptions. It's about matching your protein's biology to the host cell's capabilities.
Quick Reference Decision Tree:
Key Principles:
Start simple (E. coli) unless you have a specific reason not to
Know your protein's PTM requirements before you start (use UniProt + prediction tools)
Don't waste time on systems that fundamentally can't deliver what you need
Move up the ladder only when necessary—each step increases cost and time
Cost and speed matter—use the simplest system that works
PTMs aren't optional decorations. They're functional requirements. Choose your expression system accordingly.
Let Orbion Predict PTMs and Choose Your Expression System
Manually checking UniProt, running prediction servers, cross-referencing literature—this takes hours per protein. Worse, experimental databases have significant gaps: most proteins lack comprehensive PTM annotations because they haven't been systematically studied.
Orbion solves both problems:
1. Higher-Quality PTM Prediction
Orbion's AI models don't just look up what's in databases—they predict the complete PTM landscape from sequence and structure, even when experimental data is absent.
✓ N- and O-glycosylation sites - comprehensive prediction beyond consensus motifs
✓ Phosphorylation sites - Ser/Thr/Tyr with confidence scores
✓ Lipidation - palmitoylation, myristoylation, prenylation
✓ Disulfide bonds - predicted from structural context and evolutionary conservation
✓ Topology - membrane regions, signal peptides, domain boundaries
✓ Binding sites - active sites, allosteric sites, protein-protein interfaces
Why this matters: UniProt might show 2 experimentally validated glycosylation sites. Orbion predicts 5 additional sites that haven't been experimentally characterized yet—but will affect your expression strategy.
2. Automated Expression System Recommendation
Based on the complete PTM profile, Orbion automatically recommends the optimal expression system:
Detects glycosylation requirements → rules out E. coli
Identifies complex disulfides → suggests yeast or higher
Recognizes membrane topology → recommends insect or mammalian
Flags therapeutic context → prioritizes CHO/HEK293
The result: Get from sequence to informed construct design in minutes, not days—with higher confidence than manual database searches.

Beta-2-adrenergic receptor protocol prediction on the Orbion platform: baculovirus, Sf9 suspension, a stabilizing ligand, and a carefully engineered construct. The expression system isn’t just ‘insect’—it’s an entire strategy built around the receptor’s PTMs, folding, and stability.

