#ProteinWrapped: Create Your Holiday Card →
Characterize. Engineer. Execute.
AI-powered platform for comprehensive protein characterization, rational engineering, and experimental protocol generation—from simple proteins to multi-chain complexes.
Trusted by








#ProteinWrapped →
Characterize. Engineer. Execute.
AI-powered platform for comprehensive protein characterization, rational engineering, and experimental protocol generation—from simple proteins to multi-chain complexes.
Trusted by








#ProteinWrapped: Create Your Holiday Card →
Characterize. Engineer. Execute.
AI-powered platform for comprehensive protein characterization, rational engineering, and experimental protocol generation—from simple proteins to multi-chain complexes.
Trusted by






Protein research is a bumpy road.
Getting from sequence to structure takes months of literature review and expensive trial-and-error. Each protein and complex face the same time-consuming challenges.

Weeks Per Protocol
Designing protocols requires reviewing dozens of papers and synthesizing approaches. For novel proteins, you're starting from scratch.

Costly Failed Experiments
Wrong expression system choices or missing PTMs lead to failed experiments costing €10K-50K per protein. You're guessing without knowing what works in each system.

Mutation Trial & Error
Finding stabilizing mutations requires expensive wet lab iterations. For complexes, interface optimization adds more complexity. Each cycle costs weeks and thousands.
Protein research is a bumpy road.
Getting from sequence to structure takes months of literature review and expensive trial-and-error. Each protein and complex face the same time-consuming challenges.

Weeks Per Protocol
Designing protocols requires reviewing dozens of papers and synthesizing approaches. For novel proteins, you're starting from scratch.

Costly Failed Experiments
Wrong expression system choices or missing PTMs lead to failed experiments costing €10K-50K per protein. You're guessing without knowing what works in each system.

Mutation Trial & Error
Finding stabilizing mutations requires expensive wet lab iterations. For complexes, interface optimization adds more complexity. Each cycle costs weeks and thousands.
Protein research is a bumpy road.
Getting from sequence to structure takes months of literature review and expensive trial-and-error. Each protein and complex face the same time-consuming challenges.

Weeks Per Protocol
Designing protocols requires reviewing dozens of papers and synthesizing approaches. For novel proteins, you're starting from scratch.

Costly Failed Experiments
Wrong expression system choices or missing PTMs lead to failed experiments costing €10K-50K per protein. You're guessing without knowing what works in each system.

Mutation Trial & Error
Finding stabilizing mutations requires expensive wet lab iterations. For complexes, interface optimization adds more complexity. Each cycle costs weeks and thousands.
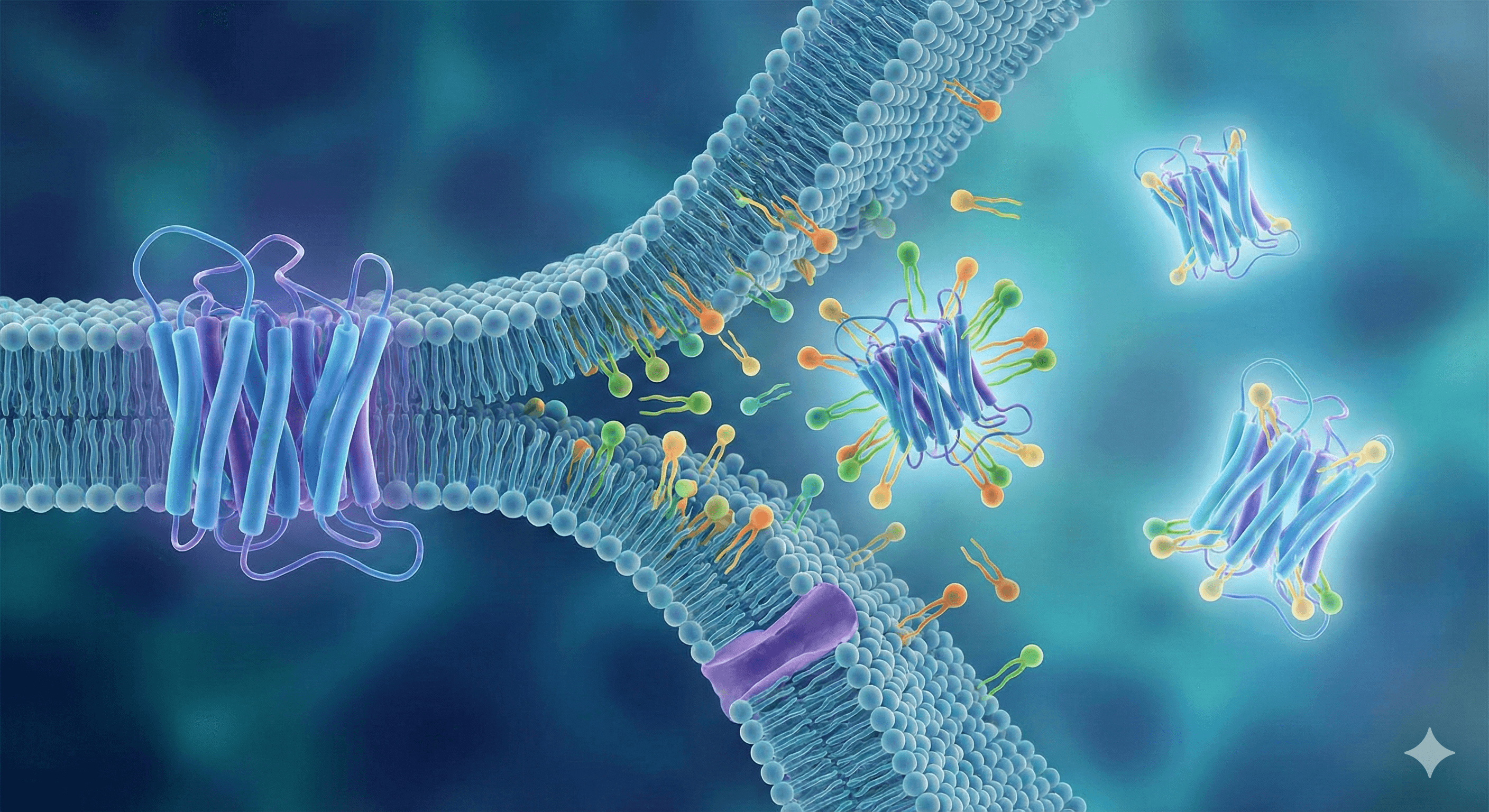
Introducing Orbion
Orbion tackles protein research challenges through AI. By combining structure and omics data, we make it easier to work on difficult proteins — from GPCRs to complexes.

Deeper Protein Understanding
Get comprehensive predictions in minutes—from PTMs and binding sites to complex interfaces and functional domains. Expression system filtering shows what PTMs will appear in E. coli, Sf9, HEK293, or yeast.

Rational Protein Engineering
Design better proteins before wet lab work—from stability mutations to construct optimization and interface engineering. All suggestions are conservation-guided to preserve function while improving expression, solubility, and crystallization.

Literature-Validated Protocols
Generate lab-ready protocols in 2 hours instead of 2 weeks—from expression and purification to crystallization and cryo-EM. Every protocol is grounded in published literature with citations.

Save 2-4 weeks per protein while preventing failed experiments through expression-specific predictions. Tackle challenging proteins — from novel GPCRs to multi-chain complexes — with confidence.
Introducing Orbion
Orbion tackles protein research challenges through AI. By combining structure and omics data, we make it easier to work on difficult proteins — from GPCRs to complexes.

Deeper Protein Understanding
Get comprehensive predictions in minutes—from PTMs and binding sites to complex interfaces and functional domains. Expression system filtering shows what PTMs will appear in E. coli, Sf9, HEK293, or yeast.

Rational Protein Engineering
Design better proteins before wet lab work—from stability mutations to construct optimization and interface engineering. All suggestions are conservation-guided to preserve function while improving expression, solubility, and crystallization.

Literature-Validated Protocols
Generate lab-ready protocols in 2 hours instead of 2 weeks—from expression and purification to crystallization and cryo-EM. Every protocol is grounded in published literature with citations.

Save 2-4 weeks per protein while preventing failed experiments through expression-specific predictions. Tackle challenging proteins — from novel GPCRs to multi-chain complexes — with confidence.
Introducing Orbion
Orbion tackles protein research challenges through AI. By combining structure and omics data, we make it easier to work on difficult proteins — from GPCRs to complexes.

Deeper Protein Understanding
Get comprehensive predictions in minutes—from PTMs and binding sites to complex interfaces and functional domains. Expression system filtering shows what PTMs will appear in E. coli, Sf9, HEK293, or yeast.

Rational Protein Engineering
Design better proteins before wet lab work—from stability mutations to construct optimization and interface engineering. All suggestions are conservation-guided to preserve function while improving expression, solubility, and crystallization.

Literature-Validated Protocols
Generate lab-ready protocols in 2 hours instead of 2 weeks—from expression and purification to crystallization and cryo-EM. Every protocol is grounded in published literature with citations.

Save 2-4 weeks per protein while preventing failed experiments through expression-specific predictions. Tackle challenging proteins — from novel GPCRs to multi-chain complexes — with confidence.
See Product in Action
Explore the platform's three main modules: comprehensive protein characterization, evaluating and generating mutants for stability and solubility, and automated protocol generation for bench work.


Characterize: Protein Intelligence Engine
Characterize: Protein Intelligence Engine
Map PTMs, binding pockets, and functional domains in minutes.
Map PTMs, binding pockets, and functional domains in minutes.


Bench: Wet-lab Co-pilot
Bench: Wet-lab Co-pilot
Predict experimental conditions and generate editable protocols.
Predict experimental conditions and generate editable protocols.



FAQ
What is Orbion?
Orbion is an AI-powered platform that helps protein researchers accelerate their work by providing comprehensive protein characterization, mutation design suggestions, and automated experimental protocol generation—all from a single protein sequence.
How accurate is Orbion's protein characterization?
Orbion achieves 90%+ accuracy for PTM predictions on well-characterized proteins when compared to experimental data. Structure predictions use AlphaFold (Monomer and Multimer) with confidence scoring via pLDDT and PAE metrics. All predictions include confidence scores so you know which regions to trust.
How are Orbion's predictions validated?
Our PTM predictions achieve 90%+ accuracy on well-characterized proteins when compared to experimental data. Protocols are grounded in published literature and cite specific papers. We're actively building wet-lab validation partnerships to further improve accuracy.
Which proteins can Orbion work with?
Orbion works with any protein sequence—cytoplasmic, membrane, GPCRs, transporters, enzymes, antibodies, and multi-chain complexes. Submit single chains or entire complexes with multiple subunits. If you have a sequence or UniProt ID, Orbion can analyze it.
Can Orbion analyze protein complexes?
Yes! Orbion supports multi-chain protein analysis using AlphaFold Multimer. Submit multiple chains to get interface predictions, chain interaction analysis, PTMs at interfaces, and complex-specific protocols. We identify which residues are at interfaces, predict interaction confidence, and generate protocols for complex assembly and characterization.
Is there a free trial?
Yes! You can have a one week free trial free to see Orbion's predictions and protocol generation in action.
How is this different from just using AlphaFold?
AlphaFold predicts structure (monomers and multimers). Orbion goes further by predicting PTMs, binding sites, interface residues for complexes, suggesting mutations, and generating complete experimental protocols—all integrated into one platform. We also provide expression system-specific predictions, conservation analysis, and complex-specific protocols not available elsewhere. For multi-chain complexes, we analyze interfaces, identify interaction hotspots, and generate assembly protocols.
What types of proteins work best with Orbion?
All protein types work with Orbion: cytoplasmic proteins, membrane proteins, GPCRs, transporters, enzymes, kinases, antibodies, and multi-chain complexes. Both well-characterized proteins and novel targets benefit from AI predictions. Membrane proteins and GPCRs especially benefit from expression system recommendations.
FAQ
What is Orbion?
Orbion is an AI-powered platform that helps protein researchers accelerate their work by providing comprehensive protein characterization, mutation design suggestions, and automated experimental protocol generation—all from a single protein sequence.
How accurate is Orbion's protein characterization?
Orbion achieves 90%+ accuracy for PTM predictions on well-characterized proteins when compared to experimental data. Structure predictions use AlphaFold (Monomer and Multimer) with confidence scoring via pLDDT and PAE metrics. All predictions include confidence scores so you know which regions to trust.
How are Orbion's predictions validated?
Our PTM predictions achieve 90%+ accuracy on well-characterized proteins when compared to experimental data. Protocols are grounded in published literature and cite specific papers. We're actively building wet-lab validation partnerships to further improve accuracy.
Which proteins can Orbion work with?
Orbion works with any protein sequence—cytoplasmic, membrane, GPCRs, transporters, enzymes, antibodies, and multi-chain complexes. Submit single chains or entire complexes with multiple subunits. If you have a sequence or UniProt ID, Orbion can analyze it.
Can Orbion analyze protein complexes?
Yes! Orbion supports multi-chain protein analysis using AlphaFold Multimer. Submit multiple chains to get interface predictions, chain interaction analysis, PTMs at interfaces, and complex-specific protocols. We identify which residues are at interfaces, predict interaction confidence, and generate protocols for complex assembly and characterization.
Is there a free trial?
Yes! You can have a one week free trial free to see Orbion's predictions and protocol generation in action.
How is this different from just using AlphaFold?
AlphaFold predicts structure (monomers and multimers). Orbion goes further by predicting PTMs, binding sites, interface residues for complexes, suggesting mutations, and generating complete experimental protocols—all integrated into one platform. We also provide expression system-specific predictions, conservation analysis, and complex-specific protocols not available elsewhere. For multi-chain complexes, we analyze interfaces, identify interaction hotspots, and generate assembly protocols.
What types of proteins work best with Orbion?
All protein types work with Orbion: cytoplasmic proteins, membrane proteins, GPCRs, transporters, enzymes, kinases, antibodies, and multi-chain complexes. Both well-characterized proteins and novel targets benefit from AI predictions. Membrane proteins and GPCRs especially benefit from expression system recommendations.
FAQ
What is Orbion?
Orbion is an AI-powered platform that helps protein researchers accelerate their work by providing comprehensive protein characterization, mutation design suggestions, and automated experimental protocol generation—all from a single protein sequence.
How accurate is Orbion's protein characterization?
Orbion achieves 90%+ accuracy for PTM predictions on well-characterized proteins when compared to experimental data. Structure predictions use AlphaFold (Monomer and Multimer) with confidence scoring via pLDDT and PAE metrics. All predictions include confidence scores so you know which regions to trust.
How are Orbion's predictions validated?
Our PTM predictions achieve 90%+ accuracy on well-characterized proteins when compared to experimental data. Protocols are grounded in published literature and cite specific papers. We're actively building wet-lab validation partnerships to further improve accuracy.
Which proteins can Orbion work with?
Orbion works with any protein sequence—cytoplasmic, membrane, GPCRs, transporters, enzymes, antibodies, and multi-chain complexes. Submit single chains or entire complexes with multiple subunits. If you have a sequence or UniProt ID, Orbion can analyze it.
Can Orbion analyze protein complexes?
Yes! Orbion supports multi-chain protein analysis using AlphaFold Multimer. Submit multiple chains to get interface predictions, chain interaction analysis, PTMs at interfaces, and complex-specific protocols. We identify which residues are at interfaces, predict interaction confidence, and generate protocols for complex assembly and characterization.
Is there a free trial?
Yes! You can have a one week free trial free to see Orbion's predictions and protocol generation in action.
How is this different from just using AlphaFold?
AlphaFold predicts structure (monomers and multimers). Orbion goes further by predicting PTMs, binding sites, interface residues for complexes, suggesting mutations, and generating complete experimental protocols—all integrated into one platform. We also provide expression system-specific predictions, conservation analysis, and complex-specific protocols not available elsewhere. For multi-chain complexes, we analyze interfaces, identify interaction hotspots, and generate assembly protocols.
What types of proteins work best with Orbion?
All protein types work with Orbion: cytoplasmic proteins, membrane proteins, GPCRs, transporters, enzymes, kinases, antibodies, and multi-chain complexes. Both well-characterized proteins and novel targets benefit from AI predictions. Membrane proteins and GPCRs especially benefit from expression system recommendations.
See Orbion's models in action.
Curious how Orbion's proprietary models streamline and accelerate protein research? Book a demo, and let us showcase you how to harness Orbion's capabilities for your research.
See Orbion's models in action.
Curious how Orbion's proprietary models streamline and accelerate protein research? Book a demo, and let us showcase you how to harness Orbion's capabilities for your research.
See Orbion's models in action.
Curious how Orbion's proprietary models streamline and accelerate protein research? Book a demo, and let us showcase you how to harness Orbion's capabilities for your research.
Orbion GmbH © Copyright 2025
Orbion GmbH © Copyright 2025
Orbion GmbH © Copyright 2025